中国物理B ›› 2020, Vol. 29 ›› Issue (10): 108707-.doi: 10.1088/1674-1056/abb659
Daiwen Sun(孙黛雯)1, Shijie Liu(刘世婕)1, Xinqi Gong(龚新奇)1,2,†( )
)
-
收稿日期:2020-06-29修回日期:2020-08-31接受日期:2020-09-09出版日期:2020-10-05发布日期:2020-10-05 -
通讯作者:Xinqi Gong(龚新奇)
Review of multimer protein–protein interaction complex topology and structure prediction
Daiwen Sun(孙黛雯)1, Shijie Liu(刘世婕)1, and Xinqi Gong(龚新奇)1,2,†
- 1
Mathematics Intelligence Application Laboratory, Institute for Mathematical Sciences, Renmin University of China , Beijing 100872,China
2Beijing Advanced Innovation Center for Structural Biology, Tshinghua University , Beijing 100094,China
-
Received:2020-06-29Revised:2020-08-31Accepted:2020-09-09Online:2020-10-05Published:2020-10-05 -
Contact:†Corresponding author. E-mail:xinqigong@ruc.edu.cn -
About author:†Corresponding author. E-mail: xinqigong@ruc.edu.cn* Project supported by the National Natural Science Foundation of China (Grant No. 31670725).
中图分类号: (Protein-protein interactions)
- 87.15.km
引用本文
Daiwen Sun(孙黛雯), Shijie Liu(刘世婕), Xinqi Gong(龚新奇). [J]. 中国物理B, 2020, 29(10): 108707-.
Daiwen Sun(孙黛雯), Shijie Liu(刘世婕), and Xinqi Gong(龚新奇)†. Review of multimer protein–protein interaction complex topology and structure prediction[J]. Chin. Phys. B, 2020, 29(10): 108707-.
"
| Optimization algorithms | Programs |
|---|---|
| Fast Fourier transformation | ZDOCK; GRAMM; DOT; SmoothDock; ClusPro; MolFit; FTDock; 3D-Dock; PIPER; pyDock; HDOCK; SDOCK; HEX; FRODOCK; InterEvDock; MDockPP; CoDockPP; HSYMDOCK; SAM |
| Monte Carlo | RosettaDock; ICM-DOCK; HADDOCK; ATTRACT |
| Genetic algorithm | DARWIN; Multi-LZerD; AutoDock |
"
| Methods | Description | Advantages | Limits | ||
|---|---|---|---|---|---|
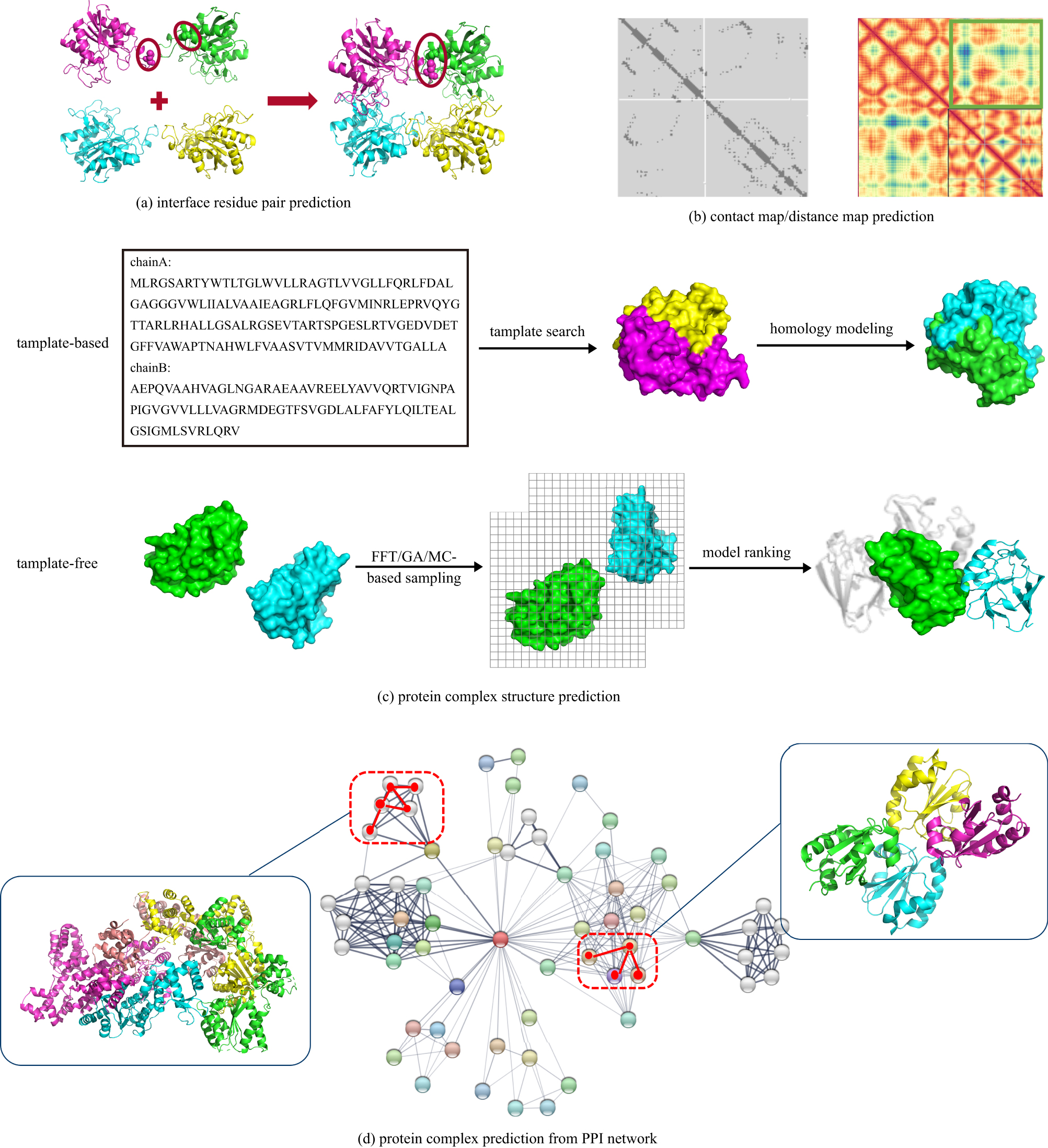
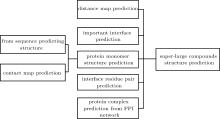
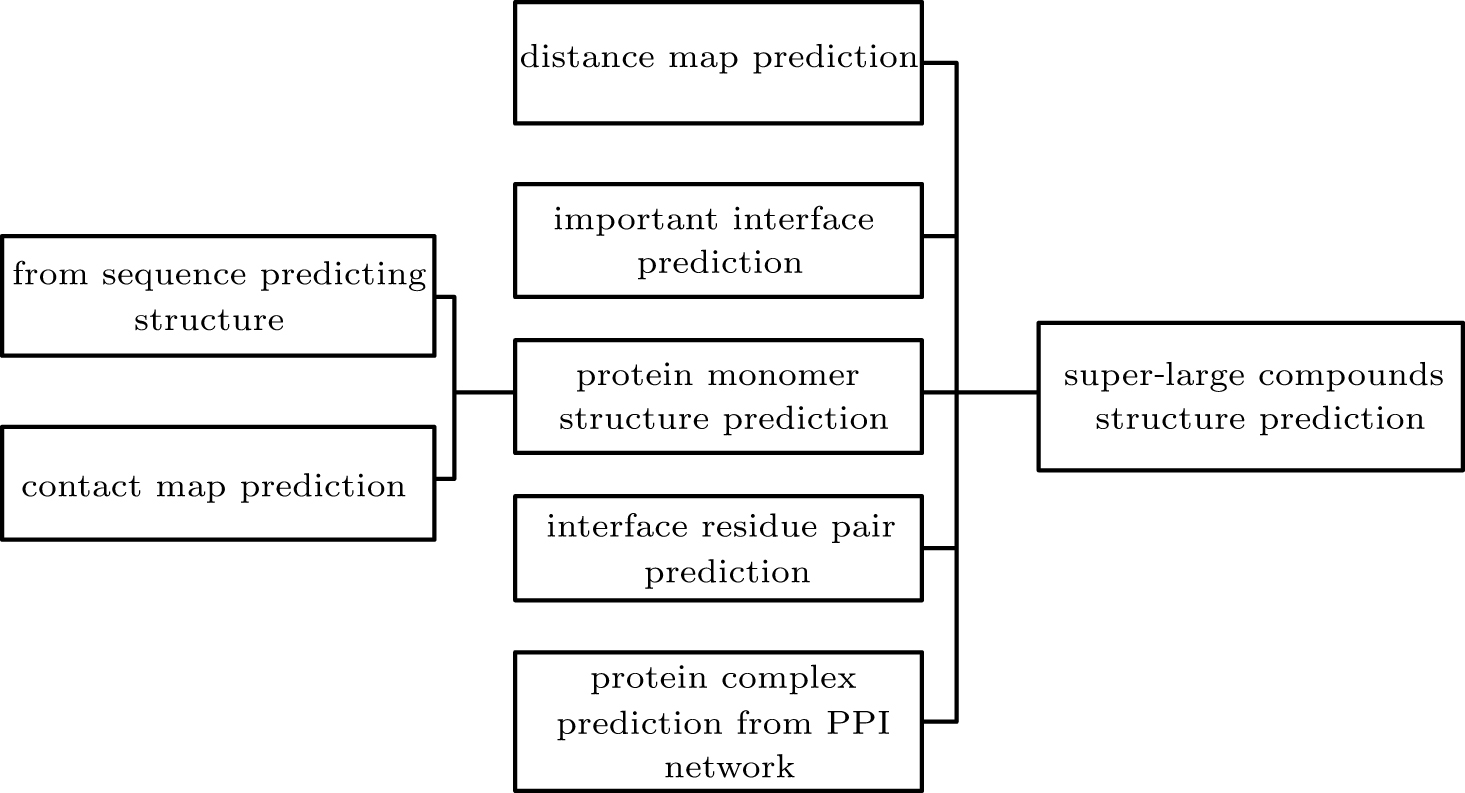
| Interface residue pair prediction | ComplexContact; RaptorX-Contact; RaptorX-Property; Gremlin; DNCON2; PSICOV; FreeContact; LSTM; LSTM with Graph Representation | Direct evolutionary coupling analysis (DCA), machine learning and deep learning methods | Interfacial residue pair prediction can help subsequent protein complex structure predictions, such as docking.Protein contact map prediction can help reconstruct the three-dimensional structure of protein complexes. | The accuracy of interface residues for prediction needs to be improved. | |
| Protein structure prediction | Template-free | ZDOCK; GRAMM; DOT; SmoothDock; ClusPro; MolFit; FTDock; 3D-Dock; PIPER; pyDock; HDOCK; SDOCK; HEX; FRODOCK; InterEvDock; MDockPP; CoDockPP; HSYMDOCK; SAM; RosettaDock; ICM-DOCK; HADDOCK; ATTRACT; DARWIN; Multi-LZerD; AutoDock | The search strategies of these methods are mainly FFT, GA and MC. | Protein docking can give all possible complex structures, some of which can also dock Cn and Dn complexes. | Designing an effective scoring function to sort the docking structure remains to be further explored. |
| Template-based | InterPreTS; Multimeric threading approach; M-Tasser; PISA; ProtCID | Using sequence or structure similarity to model protein complexes with known structures. | Template-based methods mainly reduce the possible structure by restricting the direction of protein binding. This method is more efficient than docking and can be applied to larger-scale protein complex prediction. | For proteins without a template, the structure of the complex cannot be predicted. | |
| Protein complex prediction from PPI networks | Complex prediction based on PPI network clustering | MCODE; MCL; SPC; LCMA; SuperComplex; BN; CFinder; DPClus; IPCA; CMC; ClusterONE; HACO | The protein complex is part of a known PPI network, that is, the graph composed of protein complexes and their interactions is a subgraph of the PPI network. | Some of these methods only use the PPI network for clustering, and some use additional biological information, including structure, function, organization and co-evolution infornation, etc. | The proteins that may form complexes can only be picked out from the existing PPI network. |
| Complex interaction link prediction from PPI network | GGA; HAC; ECT; RWR; MDS; Link-weighted PPI | Methods to predict actual links in the network include public neighbors-based methods and distance-based methods. | This type of method predicts possible protein--protein interactions based on existing network information. | Public neighbors-based methods have limited effect on sparse networks. |
| [1] |
Janin J 2010 Molecular bioSystems 6 2351 DOI: 10.1039/c005060c
|
| [2] |
Sudha G, Nussinov R, Srinivasan N 2014 Prog. Biophys. Mol. Biol. 116 141 DOI: 10.1016/j.pbiomolbio.2014.07.004
|
| [3] |
Zhou T M, Wang S, Xu J 2018 bioRxiv DOI: 10.1101/240754v3
|
| [4] |
Zeng H, Wang S, Zhou T, Zhao F, Li X, Wu Q, Xu J 2018 Nucleic Acids Research 46 W432 DOI: 10.1093/nar/gky420
|
| [5] |
Ching T, Himmelstein D S, Beaulieu-Jones B K et al. 2018 J. R. Soc. Interface 15 20170387 DOI: 10.1098/rsif.2017.0387
|
| [6] |
Wang S, Sun S, Li Z, Zhang R, Xu J 2017 PLoS Comput. Biol. 13 e1005324 DOI: 10.1371/journal.pcbi.1005324
|
| [7] |
Wang S, Li W, Liu S, Xu J 2016 Nucleic Acids Research 44 W430 DOI: 10.1093/nar/gkw306
|
| [8] |
Kamisetty H, Ovchinnikov S, Baker D 2013 Proc. Natl. Acad. Sci. USA 110 15674 DOI: 10.1073/pnas.1314045110
|
| [9] |
Balakrishnan S, Kamisetty H, Carbonell J G, Lee S I, Langmead C J 2011 Proteins 79 1061 DOI: 10.1002/prot.22934
|
| [10] |
Ovchinnikov S, Kamisetty H, Baker D 2014 Elife 3 e02030 DOI: 10.7554/eLife.02030.002
|
| [11] |
Adhikari B, Hou J, Cheng J 2018 Bioinformatics 34 1466 DOI: 10.1093/bioinformatics/btx781
|
| [12] |
Jones D T, Buchan D W, Cozzetto D, Pontil M 2012 Bioinformatics 28 184 DOI: 10.1093/bioinformatics/btr638
|
| [13] |
Kaján L, Hopf T A, Kalaš M, Marks D S, Rost B 2014 BMC Bioinformatics 15 85 DOI: 10.1186/1471-2105-15-85
|
| [14] |
Wang S, Li W, Zhang R, Liu S, Xu J 2016 Nucleic Acids Res. 44 W361 DOI: 10.1093/nar/gkw307
|
| [15] |
Xu G, Wang Q, Ma J 2020 Journal of Chemical Theory and Computation 16 3970 DOI: 10.1021/acs.jctc.0c00186
|
| [16] |
Zhao Z, Gong X 2019 IEEEACM Transactions on Computational Biology and Bioinformatics 16 1753 DOI: 10.1109/TCBB.2017.2706682
|
| [17] |
Sun D, Gong X 2020 Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 140504 DOI: 10.1016/j.bbapap.2020.140504
|
| [18] |
Liu J, Gong X 2019 BMC Bioinformatics 20 609 DOI: 10.1186/s12859-019-3199-1
|
| [19] |
Vreven T, Hwang H, Pierce B G, Weng Z 2014 Brief Bioinform 15 169 DOI: 10.1093/bib/bbt047
|
| [20] |
Zhang Q, Feng T, Xu L, Sun H, Pan P, Li Y, Li D, Hou T 2016 Curr. Drug Targets 17 1586 DOI: 10.2174/1389450117666160112112640
|
| [21] |
Ritchie D W, Kemp G J 2000 Proteins: Structure, Function, and Bioinformatics 39 178 DOI: 10.1002/(ISSN)1097-0134
|
| [22] |
Lee K, Czaplewski C, Kim S Y, Lee J 2005 Journal of Computational Chemistry 26 78 DOI: 10.1002/(ISSN)1096-987X
|
| [23] |
Ritchie D W 2003 Proteins: Structure, Function, and Bioinformatics 52 98 DOI: 10.1002/(ISSN)1097-0134
|
| [24] |
Pierce B, Tong W, Weng Z 2005 Bioinformatics 21 1472 DOI: 10.1093/bioinformatics/bti229
|
| [25] |
Chen R, Li L, Weng Z 2003 Proteins 52 80 DOI: 10.1002/(ISSN)1097-0134
|
| [26] |
André I, Bradley P, Wang C, Baker D 2007 Proc. Natl. Acad. Sci. USA 104 17656 DOI: 10.1073/pnas.0702626104
|
| [27] |
Comeau S R, Gatchell D W, Vajda S, Camacho C J 2004 Bioinformatics 20 45 DOI: 10.1093/bioinformatics/btg371
|
| [28] |
Yan Y, Tao H, He J, Huang S Y 2020 Nature Protocols 15 1829 DOI: 10.1038/s41596-020-0312-x
|
| [29] |
Yan Y, Zhang D, Zhou P, Li B, Huang S Y 2017 Nucleic Acids Res. 45 W365 DOI: 10.1093/nar/gkx407
|
| [30] |
Yan Y, Tao H, Huang S Y 2018 Nucleic Acids Res. 46 W423 DOI: 10.1093/nar/gky398
|
| [31] |
Ritchie D W, Grudinin S 2016 Journal of Applied Crystallography 49 158 DOI: 10.1107/S1600576715022931
|
| [32] |
Yu H, Luscombe N M, Lu H X, Zhu X, Xia Y, Han J D J, Bertin N, Chung S, Vidal M, Gerstein M 2004 Genome Research 14 1107 DOI: 10.1101/gr.1774904
|
| [33] |
Aloy P, Russell R B 2002 Proc. Natl. Acad. Sci. USA 99 5896 DOI: 10.1073/pnas.092147999
|
| [34] |
Aloy P, Russell R B 2003 Bioinformatics 19 161 DOI: 10.1093/bioinformatics/19.1.161
|
| [35] |
Lu L, Lu H, Skolnick J 2002 Proteins 49 350 DOI: 10.1002/(ISSN)1097-0134
|
| [36] |
Chen H, Skolnick J 2008 Biophys J. 94 918 DOI: 10.1529/biophysj.107.114280
|
| [37] |
Launay G, Simonson T 2008 BMC Bioinformatics 9 427 DOI: 10.1186/1471-2105-9-427
|
| [38] |
Aloy P, Böttcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin A C, Bork P, Superti-Furga G, Serrano L 2004 Science 303 2026 DOI: 10.1126/science.1092645
|
| [39] |
Nye T M W, Berzuini C, Gilks W R, Babu M M, Teichmann S A 2004 Bioinformatics 21 993 DOI: 10.1093/bioinformatics/bti086
|
| [40] |
Krissinel E, Henrick K 2007 Journal of Molecular Biology 372 774 DOI: 10.1016/j.jmb.2007.05.022
|
| [41] |
Xu Q, Dunbrack R L 2010 Nucleic Acids Res. 39 D761 DOI: 10.1093/nar/gkq1059
|
| [42] |
Yu H, Braun P, Yildirim M A, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual J F, Dricot A, Vazquez A, Murray R R, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet A S, Motyl A, Hudson M E, Park J, Xin X, Cusick M E, Moore T, Boone C, Snyder M, Roth F P, Barabási A L, Tavernier J, Hill D E, Vidal M 2008 Science 322 104 DOI: 10.1126/science.1158684
|
| [43] |
Tarassov K, Messier V, Landry C R, Radinovic S, Serna Molina M M, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick S W 2008 Science 320 1465 DOI: 10.1126/science.1153878
|
| [44] |
Krogan N J, Cagney G, Yu H et al. 2006 Nature 440 637 DOI: 10.1038/nature04670
|
| [45] |
Gavin A C, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen L J, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier M A, Hoffman V, Hoefert C, Klein K, Hudak M, Michon A M, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick J M, Kuster B, Bork P, Russell R B, Superti-Furga G 2006 Nature 440 631 DOI: 10.1038/nature04532
|
| [46] |
Pržulj N 2011 Bioessays 33 115 DOI: 10.1002/bies.v33.2
|
| [47] |
Yu H, Kim P M, Sprecher E, Trifonov V, Gerstein M 2007 PLoS Comput. Biol. 3 e59 DOI: 10.1371/journal.pcbi.0030059
|
| [48] |
Jeong H, Mason S P, Barabási A L, Oltvai Z N 2001 Nature 411 41 DOI: 10.1038/35075138
|
| [49] |
Han J D J, Bertin N, Hao T, Goldberg D S, Berriz G F, Zhang L V, Dupuy D, Walhout A J M, Cusick M E, Roth F P, Vidal M 2004 Nature 430 88 DOI: 10.1038/nature02555
|
| [50] |
Wang J, Li M, Deng Y, Pan Y 2010 BMC Genomics 11 3 S10 DOI: 10.1186/1471-2164-11-s3-s10
|
| [51] |
Ulitsky I, Shamir R 2009 Bioinformatics 25 1158 DOI: 10.1093/bioinformatics/btp118
|
| [52] |
Sharan R, Ulitsky I, Shamir R 2007 Mol. Syst. Biol. 3 88 DOI: 10.1038/msb4100129
|
| [53] |
Lee K, Chuang H Y, Beyer A, Sung M K, Huh W K, Lee B, Ideker T 2008 Nucleic Acids Res. 36 e136 DOI: 10.1186/1471-2164-11-s3-s10
|
| [54] |
King A D, Przulj N, Jurisica I 2004 Bioinformatics 20 3013 DOI: 10.1093/bioinformatics/bth351
|
| [55] |
Friedel C C, Krumsiek J, Zimmer R 2009 J. Comput. Biol. 16 971 DOI: 10.1089/cmb.2009.0023
|
| [56] |
Chua H N, Sung W K, Wong L 2006 Bioinformatics 22 1623 DOI: 10.1093/bioinformatics/btl145
|
| [57] |
Bader G D, Hogue C W V 2002 Nature Biotechnology 20 991 DOI: 10.1038/nbt1002-991
|
| [58] |
Asthana S, King O D, Gibbons F D, Roth F P 2004 Genome Research 14 1170 DOI: 10.1101/gr.2203804
|
| [59] |
Kim Y A, Wuchty S, Przytycka T M 2011 PLoS Comput. Biol. 7 e1001095 DOI: 10.1371/journal.pcbi.1001095
|
| [60] |
Ideker T, Sharan R 2008 Genome Research 18 644 DOI: 10.1101/gr.071852.107
|
| [61] |
Hidalgo C A, Blumm N, Barabási A L, Christakis N A 2009 PLoS Comput. Biol. 5 e1000353
|
| [62] |
Hannum G, Srivas R, Guénolé A, van Attikum H, Krogan N J, Karp R M, Ideker T 2009 PLoS Genet. 5 e1000782
|
| [63] |
Chuang H Y, Lee E, Liu Y T, Lee D, Ideker T 2007 Molecular Systems Biology 3 140 DOI: 10.1038/msb4100180
|
| [64] |
Huang H, Jedynak B M, Bader J S 2007 PLoS Comput. Biol. 3 e214 DOI: 10.1371/journal.pcbi.0030214
|
| [65] |
Lei C, Ruan J 2012 IEEE International Conference on Bioinformatics and Biomedicine 4–7 October, 2012 1 6 https://ieeexplore.ieee.org/document/6392693/
|
| [66] |
Srihari S, Yong C H, Wong L 2017 Computational prediction of protein complexes from protein interaction networks Association for Computing Machinery DOI: 10.1145/3064650
|
| [67] |
Bader G D, Hogue C W 2003 BMC bioinformatics 4 2 DOI: 10.1186/1471-2105-4-2
|
| [68] |
Pereira Leal J B, Enright A J, Ouzounis C A 2004 PROTEINS: Structure, Function, and Bioinformatics 54 49 DOI: 10.1002/prot.10505
|
| [69] |
Brohee S, Van Helden J 2006 BMC Bioinformatics 7 488 DOI: 10.1186/1471-2105-7-488
|
| [70] |
Pu S, Vlasblom J, Emili A, Greenblatt J, Wodak S J 2007 Proteomics 7 944 DOI: 10.1002/(ISSN)1615-9861
|
| [71] |
Blatt M, Wiseman S, Domany E 1996 Phys. Rev. Lett. 76 3251 DOI: 10.1103/PhysRevLett.76.3251
|
| [72] |
Getz G, Vendruscolo M, Sachs D, Domany E 2002 Proteins: Structure, Function, and Bioinformatics 46 405 DOI: 10.1002/(ISSN)1097-0134
|
| [73] |
Getz G, Levine E, Domany E 2000 Proc. Natl. Acad. Sci. USA 97 12079 DOI: 10.1073/pnas.210134797
|
| [74] |
Spirin V, Mirny L A 2003 Proc. Natl. Acad. Sci. USA 100 12123 DOI: 10.1073/pnas.2032324100
|
| [75] |
Li X L, Foo C S, Tan S H, Ng S K 2005 Genome Informatics 16 260 DOI: 10.11234/gi1990.16.2_260
|
| [76] |
Qi Y, Balem F, Faloutsos C, Klein-Seetharaman J, Bar-Joseph Z 2008 Bioinformatics 24 i250 DOI: 10.1093/bioinformatics/btn164
|
| [77] |
Yong C H, Liu G, Chua H N, Wong L BMC Systems Biology S13 DOI: 10.1186/1752-0509-6-S2-S13
|
| [78] |
Srihari S, Leong H W 2012 International Journal of Bioinformatics Research and Applications 8 286 DOI: 10.1504/IJBRA.2012.048962
|
| [79] |
Adamcsek B, Palla G, Farkas I J, Derenyi I, Vicsek T 2006 Bioinformatics 22 1021 DOI: 10.1093/bioinformatics/btl039
|
| [80] |
Altaf-Ul-Amin M, Shinbo Y, Mihara K, Kurokawa K, Kanaya S 2006 BMC Bioinformatics 7 207 DOI: 10.1186/1471-2105-7-207
|
| [81] |
Li M, Chen J E, Wang J X, Hu B, Chen G 2008 BMC Bioinformatics 9 398 DOI: 10.1186/1471-2105-9-398
|
| [82] |
Liu G, Wong L, Chua H N 2009 Bioinformatics 25 1891 DOI: 10.1093/bioinformatics/btp311
|
| [83] |
Nepusz T, Yu H, Paccanaro A 2012 Nat. Methods 9 471 DOI: 10.1038/NMETH.1938
|
| [84] |
Wang H, Kakaradov B, Collins S R, Karotki L, Fiedler D, Shales M, Shokat K M, Walther T C, Krogan N J, Koller D 2009 Mol. Cell Proteomics 8 1361 DOI: 10.1074/mcp.M800490-MCP200
|
| [85] |
Tong H, Faloutsos C, Pan J Y Sixth international conference on data mining (ICDM’06) 613 622 http://www.cs.cmu.edu/~christos/PUBLICATIONS/icdm06-rwr.pdf
|
| [86] |
Radicchi F, Castellano C, Cecconi F, Loreto V, Parisi D 2004 Proc. Natl. Acad. Sci. USA 101 2658 DOI: 10.1073/pnas.0400054101
|
| [87] |
Li A, Horvath S 2007 Bioinformatics 23 222 DOI: 10.1093/bioinformatics/btl581
|
| [88] |
Fouss F, Pirotte A, Renders J, Saerens M 2007 IEEE Transactions on Knowledge and Data Engineering 19 355 DOI: 10.1109/TKDE.2007.46
|
| [89] |
Lü L, Zhou T 2011 Physica A 390 1150 DOI: 10.1016/j.physa.2010.11.027
|
| [90] |
Wang C, Ding C, Yang Q, Holbrook S R 2007 Genome Biol. 8 R271 DOI: 10.1186/gb-2007-8-12-r271
|
| [91] |
Fang Y, Benjamin W, Sun M, Ramani K 2011 PLoS One 6 e19349 DOI: 10.1371/journal.pone.0019349
|
| [92] |
Xu Q, Xiang E W, Yang Q 2011 Proteomics 11 3818 DOI: 10.1002/pmic.201100146
|
| [93] |
Park Y, Bader J S 2011 BMC Bioinformatics 12 S44 DOI: 10.1186/1471-2105-12-S1-S44
|
| [94] |
Kuchaiev O, Rasajski M, Higham D J, Przulj N 2009 PLoS Comput. Biol. 5 e1000454 DOI: 10.1371/journal.pcbi.1000454
|
| [95] |
Lei C, Ruan J 2012 Bioinformatics 29 355 DOI: 10.1093/bioinformatics/bts688
|
| [96] |
Wang L, Hu K, Tang Y 2014 Current Bioinformatics 9 246 DOI: 10.2174/1574893609666140516005740
|
| [97] |
Bartoli L, Capriotti E, Fariselli P, Martelli P L, Casadio R 2008 Methods Mol. Biol. 413 199 DOI: 10.1371/journal.pcbi.1000454
|
| [98] |
Vassura M, Margara L, Di Lena P, Medri F, Fariselli P, Casadio R 2008 IEEEACM Trans. Comput. Biol. Bioinform. 5 357 DOI: 10.1109/tcbb.2008.27
|
| [99] |
Vendruscolo M, Kussell E, Domany E 1997 Fold Des. 2 295 DOI: 10.1016/S1359-0278(97)00041-2
|
| [100] |
Breu H, Kirkpatrick D G 1998 Computational Geometry 9 3 DOI: 10.1016/S0925-7721(97)00014-X
|
| [101] |
Zhang C, Mortuza S M, He B, Wang Y, Zhang Y 2018 Proteins 86 1 136 DOI: 10.1002/prot.25414
|
| [102] |
Baú D, Martin A J M, Mooney C, Vullo A, Walsh I, Pollastri G 2006 BMC Bioinformatics 7 402 DOI: 10.1186/1471-2105-7-402
|
| [103] |
Kukic P, Mirabello C, Tradigo G, Walsh I, Veltri P, Pollastri G 2014 BMC Bioinformatics 15 6 DOI: 10.1186/1471-2105-15-6
|
| [104] |
Walsh I, Baù D, Martin A J M, Mooney C, Vullo A, Pollastri G 2009 BMC Structural Biology 9 5 DOI: 10.1186/1472-6807-9-5
|
| [105] |
Fariselli P, Casadio R 2001 Bioinformatics 17 957 DOI: 10.1093/bioinformatics/17.10.957
|
| [106] |
Martelli P L, Fariselli P, Malaguti L, Casadio R 2002 Protein Engineering, Design and Selection 15 951 DOI: 10.1093/protein/15.12.951
|
| [107] |
Ceroni A, Passerini A, Vullo A, Frasconi P 2006 Nucleic Acids Res. 34 W177 DOI: 10.1093/nar/gkl266
|
| [108] |
Tsai C H, Chen B J, Chan C H, Liu H L, Kao C Y 2005 Bioinformatics 21 4416 DOI: 10.1093/bioinformatics/bti715
|
| [109] |
Vullo A, Frasconi P 2004 Bioinformatics 20 653 DOI: 10.1093/bioinformatics/btg463
|
| [110] |
Ferrè F, Clote P 2005 Nucleic Acids Res 33 W230 DOI: 10.1093/nar/gki412
|
| [111] |
Schaarschmidt J, Monastyrskyy B, Kryshtafovych A, Bonvin A 2018 Proteins 86 1 51 DOI: 10.1002/prot.25407
|
| [112] |
Seemayer S, Gruber M, Söding J 2014 Bioinformatics 30 3128 DOI: 10.1093/bioinformatics/btu500
|
| [113] |
Jones D T, Kandathil S M 2018 Bioinformatics 34 3308 DOI: 10.1093/bioinformatics/bty341
|
| [114] |
Hanson J, Paliwal K, Litfin T, Yang Y, Zhou Y 2018 Bioinformatics 34 4039 DOI: 10.1093/bioinformatics/bty481
|
| [115] |
Senior A W, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson A W, Bridgland A 2019 Proteins: Structure, Function, and Bioinformatics 87 1141 DOI: 10.1002/prot.v87.12
|
| [116] |
Dong Q, Wang X, Lin L, Guan Y 2007 BMC Bioinformatics 8 147 DOI: 10.1186/1471-2105-8-147
|
| [117] |
Minhas F u A A, Geiss B J, Ben-Hur A 2014 Proteins 82 1142 DOI: 10.1002/prot.v82.7
|
| [118] |
Zellner H, Staudigel M, Trenner T, Bittkowski M, Wolowski V, Icking C, Merkl R 2012 Proteins 80 154 DOI: 10.1002/prot.23172
|
| [119] |
Wang B, Chen P, Huang D S, Li J J, Lok T M, Lyu M R 2006 FEBS Lett. 580 380 DOI: 10.1016/j.febslet.2005.11.081
|
| [120] |
Bradford J R, Westhead D R 2005 Bioinformatics 21 1487 DOI: 10.1093/bioinformatics/bti242
|
| [121] |
Koike A, Takagi T 2004 Protein Eng. Des. Sel. 17 165 DOI: 10.1093/protein/gzh020
|
| [122] |
Sriwastava B K, Basu S, Maulik U 2015 J. Biosci. 40 809 DOI: 10.1007/s12038-015-9564-y
|
| [123] |
Singh G, Dhole K, Pai P P, Mondal S 2014 SPRINGS: prediction of protein–protein interaction sites using artificial neural networks Report No. 2167–9843 DOI: 10.7287/peerj.preprints.266v2
|
| [124] |
Ofran Y, Rost B 2007 Bioinformatics 23 e13 DOI: 10.1093/bioinformatics/btl303
|
| [125] |
Chen H, Zhou H X 2005 Proteins 61 21 DOI: 10.1002/prot.20514
|
| [126] |
Ofran Y, Rost B 2003 FEBS Lett. 544 236 DOI: 10.1016/S0014-5793(03)00456-3
|
| [127] |
Fariselli P, Pazos F, Valencia A, Casadio R 2002 Eur. J. Biochem. 269 1356 DOI: 10.1046/j.1432-1033.2002.02767.x
|
| [128] |
Zhou H X, Shan Y 2001 Proteins: Structure, Function, and Bioinformatics 44 336 DOI: 10.1002/(ISSN)1097-0134
|
| [129] |
Bradford J R, Needham C J, Bulpitt A J, Westhead D R 2006 J. Mol. Biol. 362 365 DOI: 10.1016/j.jmb.2006.07.028
|
| [130] |
Neuvirth H, Raz R, Schreiber G 2004 J. Mol. Biol. 338 181 DOI: 10.1016/j.jmb.2004.02.040
|
| [131] |
Geng H, Lu T, Lin X, Liu Y, Yan F 2015 Biochemistry Research International 2015 978193 DOI: 10.1155/2015/978193
|
| [132] |
Murakami Y, Mizuguchi K 2010 Bioinformatics 26 1841 DOI: 10.1093/bioinformatics/btq302
|
| [133] |
Chen X W, Jeong J C 2009 Bioinformatics 25 585 DOI: 10.1093/bioinformatics/btp039
|
| [134] |
Northey T C, Barešić A, Martin A C R 2018 Bioinformatics 34 223 DOI: 10.1093/bioinformatics/btx585
|
| [135] |
Li B Q, Feng K Y, Chen L, Huang T, Cai Y D 2012 PLoS One 7 e43927 DOI: 10.1371/journal.pone.0043927
|
| [136] |
Sikić M, Tomić S, Vlahovicek K 2009 PLoS Comput. Biol. 5 e1000278 DOI: 10.1371/journal.pcbi.1000278
|
| [137] |
Wei Z S, Yang J Y, Shen H B, Yu D J 2015 IEEE Trans. Nanobioscience 14 746 DOI: 10.1109/TNB.2015.2475359
|
| [138] |
Li M H, Lin L, Wang X L, Liu T 2007 Bioinformatics 23 597 DOI: 10.1093/bioinformatics/btl660
|
| [139] |
Wang D D, Wang R, Yan H 2014 Neurocomputing 128 258 DOI: 10.1016/j.neucom.2012.12.062
|
| [140] |
Dhole K, Singh G, Pai P P, Mondal S 2014 J. Theor. Biol. 348 47 DOI: 10.1016/j.jtbi.2014.01.028
|
| [141] |
Jia J, Liu Z, Xiao X, Liu B, Chou K C 2016 J. Biomol. Struct. Dyn. 34 1946 DOI: 10.1080/07391102.2015.1095116
|
| [142] |
Deng L, Guan J, Dong Q, Zhou S 2009 BMC Bioinformatics 10 426 DOI: 10.1186/1471-2105-10-426
|
| [143] |
Chen P, Li J 2010 BMC Bioinformatics 11 402 DOI: 10.1186/1471-2105-11-402
|
| [144] |
Du X, Sun S, Hu C, Li X, Xia J 2016 J. Biol. Res. (Thessalon) 23 10 DOI: 10.1186/s40709-016-0046-7
|
| [145] |
Krizhevsky A, Sutskever I, Hinton G E Advances in Neural Information Processing Systems 1097 1105 DOI: 10.1186/s40709-016-0046-7
|
| [1] | . [J]. 中国物理B, 2021, 30(1): 18703-. |
| [2] | Xiang Li(李翔), Hong Qi(祁宏), Xiao-Cui Zhang(张晓翠), Fei Xu(徐飞), Zhi-Yong Yin(尹智勇), Shi-Yang Huang(黄世阳), Zhao-Shou Wang(王兆守), Jian-Wei Shuai(帅建伟). [J]. 中国物理B, 2020, 29(10): 108702-. |
| [3] | 杨为, 来鲁华. Computational design of proteins with novel structure and functions[J]. 中国物理B, 2016, 25(1): 18702-018702. |
| [4] | 毛志秀, 陈笑风, 陈彬. Stability of focal adhesion enhanced by its inner force fluctuation[J]. 中国物理B, 2015, 24(8): 88702-088702. |
| [5] | 张洪艳, 杨立泉, 孟岚, 聂家财, 宁廷银, 刘卫敏, 孙嘉宇, 汪鹏飞. [J]. 中国物理B, 2012, 21(2): 20601-020601. |
|
||