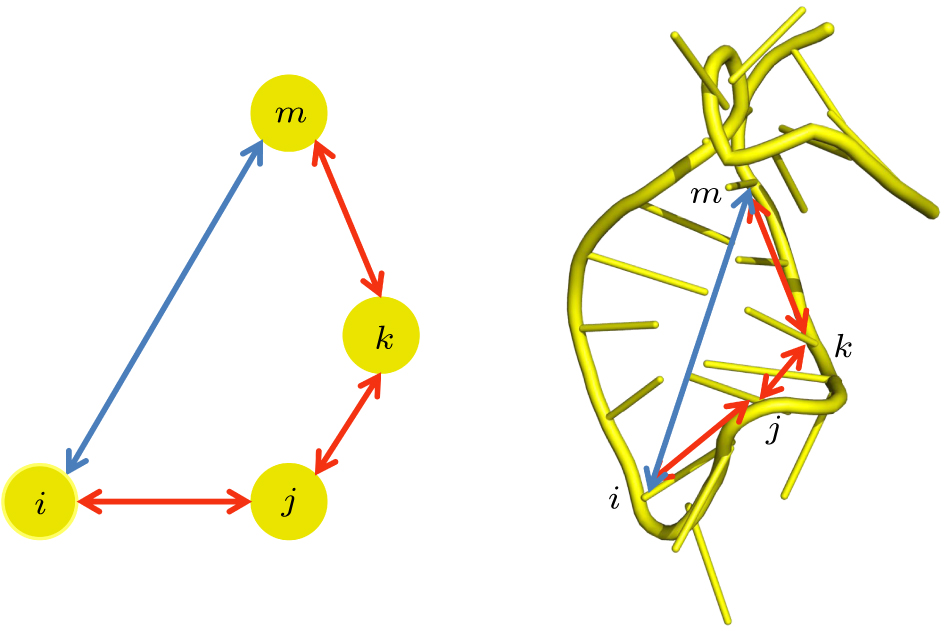
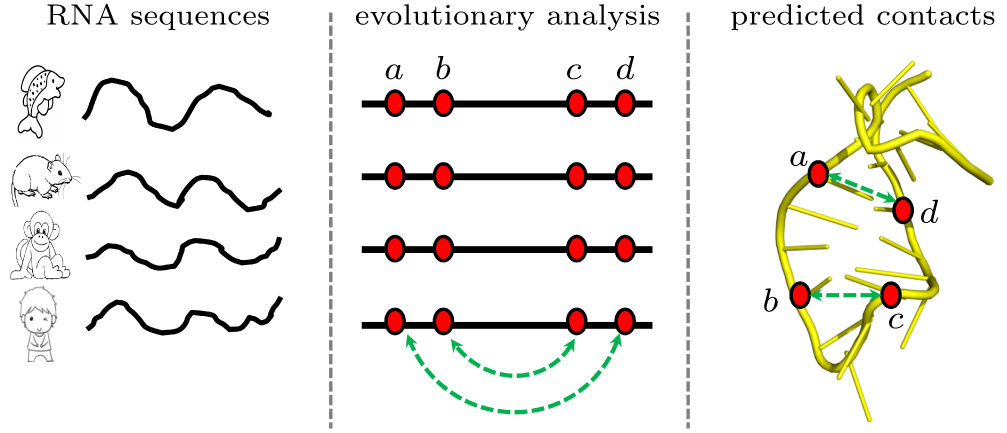
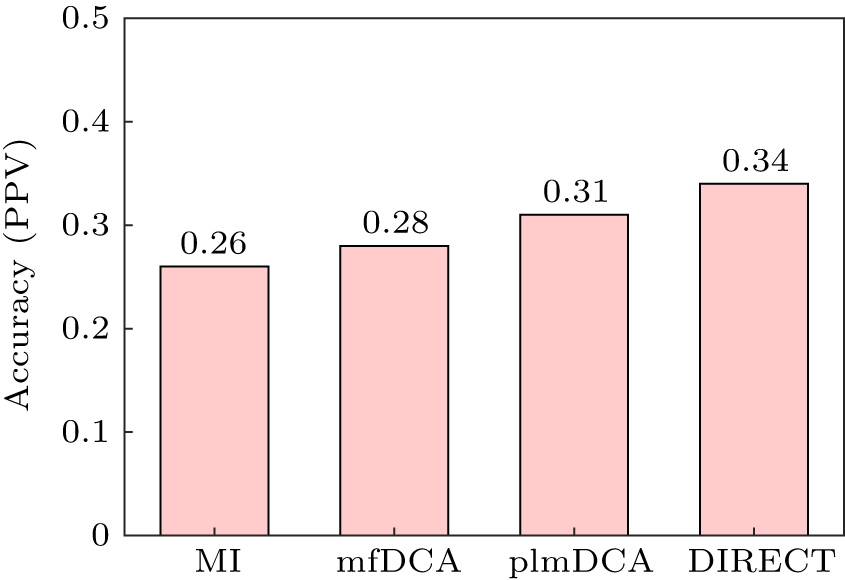
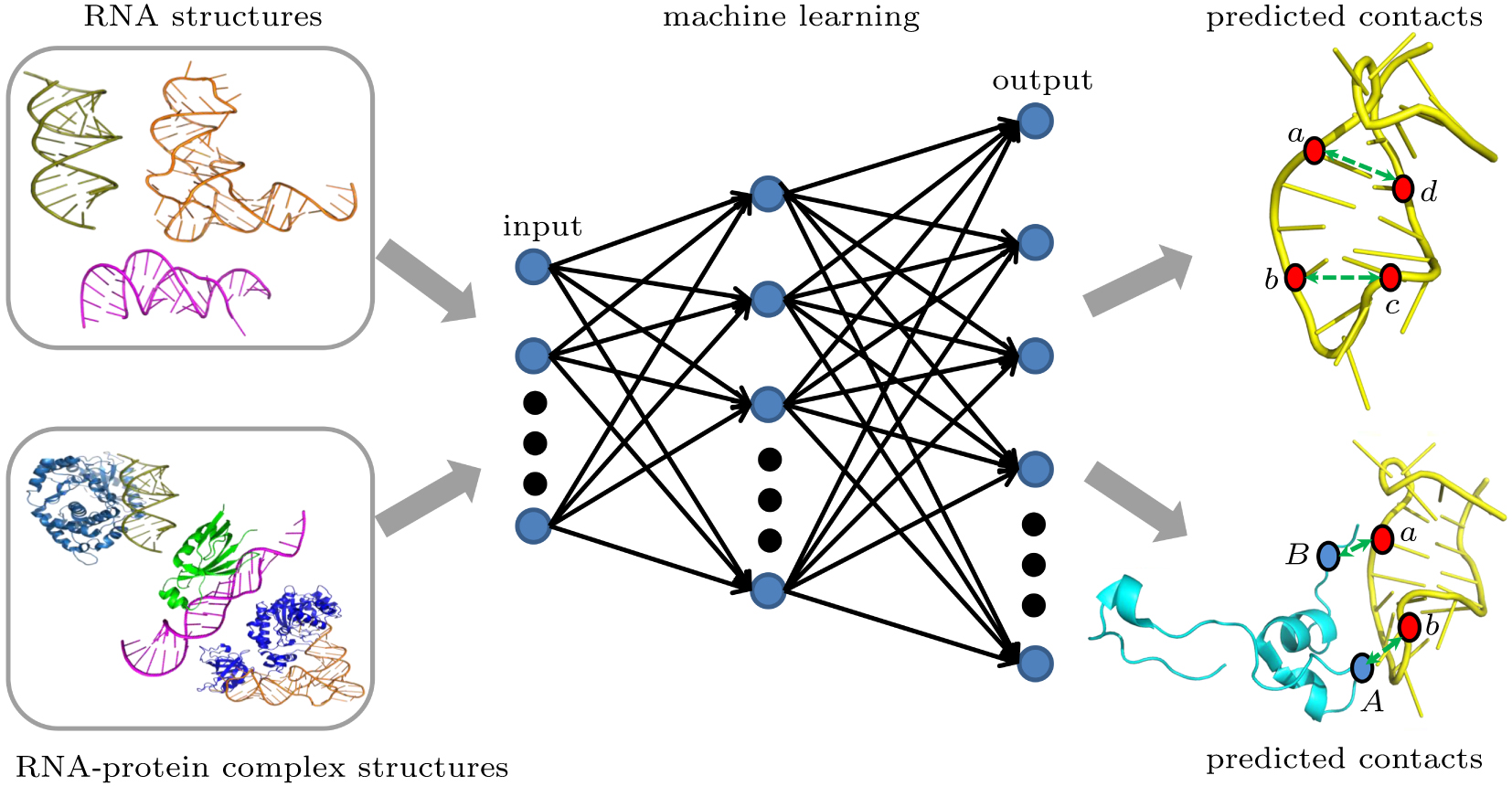
中国物理B ›› 2020, Vol. 29 ›› Issue (10): 108708-.doi: 10.1088/1674-1056/abb7f3
Huiwen Wang(王慧雯)1, Yunjie Zhao(赵蕴杰)1,†( )
)
-
收稿日期:2020-04-30修回日期:2020-07-07接受日期:2020-09-14出版日期:2020-10-05发布日期:2020-10-05 -
通讯作者:Yunjie Zhao(赵蕴杰)
Methods and applications of RNA contact prediction
Huiwen Wang(王慧雯) and Yunjie Zhao(赵蕴杰)†
- 1
Institute of Biophysics and Department of Physics, Central China Normal University , Wuhan 430079,China
-
Received:2020-04-30Revised:2020-07-07Accepted:2020-09-14Online:2020-10-05Published:2020-10-05 -
Contact:†Corresponding author. E-mail:yjzhaowh@mail.ccnu.edu.cn -
About author:†Corresponding author. E-mail: yjzhaowh@mail.ccnu.edu.cn* Project supported by the National Natural Science Foundation of China (Grant No. 11704140) and Self-determined Research Funds of CCNU from the Colleges’ Basic Research and Operation of MOE (Grant No. CCNU20TS004).
中图分类号: (RNA)
- 87.14.gn
引用本文
Huiwen Wang(王慧雯), Yunjie Zhao(赵蕴杰). [J]. 中国物理B, 2020, 29(10): 108708-.
Huiwen Wang(王慧雯) and Yunjie Zhao(赵蕴杰)†. Methods and applications of RNA contact prediction[J]. Chin. Phys. B, 2020, 29(10): 108708-.
"
| Method name | Input information | Comments | Link | Reference |
|---|---|---|---|---|
| Mutual Information | RNA sequence | intramolecular contacts | http://dca.rice.edu/portal/dca/home | [ |
| mpDCA | RNA sequence | intramolecular contacts | not available | [ |
| mfDCA | RNA sequence | intramolecular contacts | http://dca.rice.edu/portal/dca/home | [ |
| plmDCA | RNA sequence | intramolecular contacts | https://github.com/magnusekeberg/plmDCA | [ |
| DIRECT | RNA sequence and structure | intramolecular contacts | https://zhaolab.com.cn/DIRECT/ | [ |
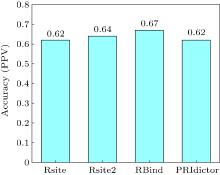
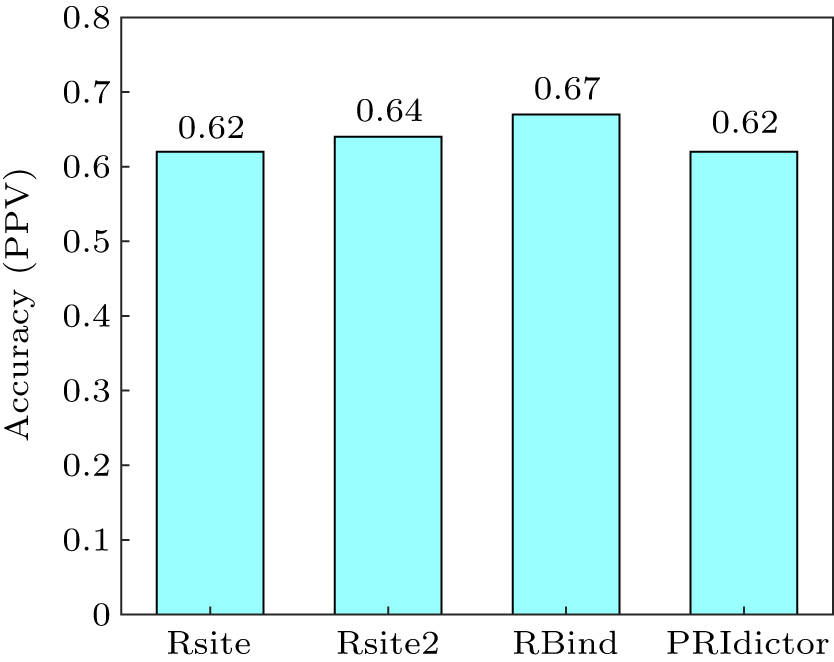
| Rsite | RNA structure | intermolecular contacts | http://www.cuilab.cn/rsite | [ |
| Rsite2 | RNA structure | intermolecular contacts | http://www.cuilab.cn/rsite2 | [ |
| RBind | RNA structure | intermolecular contacts | https://zhaolab.com.cn/RBind/ | [ |
| PRIdictor | RNA and protein sequences | intermolecular contacts | http://bclab.inha.ac.kr/pridictor/ | [ |
| [1] |
Wang J, Zhao Y, Zhu C, Xiao Y 2015 Nucleic Acids Res. 43 e63
|
| [2] |
Wang J, Mao K, Zhao Y, Zeng C, Xiang J, Zhang Y, Xiao Y 2017 Nucleic Acids Res. 45 6299
|
| [3] |
Zelinger L, Swaroop A 2018 Trends Genet. 34 341
|
| [4] |
Lu D, Thum T 2019 Nat. Rev. Cardiol. 16 661
|
| [5] |
Huang Y, Li H, Xiao Y 2018 Bioinformatics 34 1238
|
| [6] |
Zhang J, Zhang Y J, Wang W 2010 Chin. Phys. Lett. 27 118702
|
| [7] |
Nithin C, Ghosh P, Bujnicki J M 2018 Genes 9 432
|
| [8] |
Wang H, Wang K, Guan Z, Jian Y, Jia Y, Kashanchi F, Zeng C, Zhao 2017 Chin. Phys. B 26 128702
|
| [9] |
Yan Y, Huang S Y 2018 Bioinformatics 34 453
|
| [10] |
Yan Y, Zhang D, Zhou P, Li B, Huang S Y 2017 Nucleic Acids Res. 45 W365
|
| [11] |
Zhao Y, Jian Y, Liu Z, Liu H, Liu Q, Chen C, Li Z, Wang L, Huang H H, Zeng C 2017 Sci. Rep. 7 2876
|
| [12] |
Yan Y, Wen Z, Zhang D, Huang S Y 2018 Nucleic Acids Res. 46 e56
|
| [13] |
Bao L, Wang J, Xiao Y 2019 Phys. Rev. E 100 022412
|
| [14] |
Wang H, Qiu J, Liu H, Jian Y, Xu Y, Jia Y, Kashanchi F, Zeng C, Zhao Y 2019 BMC Bioinformatics 20 617
|
| [15] |
Karn J, Keen N J, Churcher M J, Aboul-ela F, Varani G, Hamy F, Felder E R, Heizmann G, Klimkait T 1998 Pharmacochemistry Library 29 121
|
| [16] |
Abulwerdi F A, Grice S F J L 2017 Curr. Pharm. Des. 23 4112
|
| [17] |
Zhao Y, Chen H, Du C, Jian Y, Li H, Xiao Y, Saifuddin M, Kashanchi F, Zeng C 2018 Int. J. Pept. Res. Ther. 25 807
|
| [18] |
Romby P, Charpentier E 2010 Cell. Mol. Life Sci. 67 217
|
| [19] |
Zhou T, Wang H, Song L, Zhao Y 2020 J. Theor. Comput. Chem. 19 2040001
|
| [20] |
Lou Y, Chen B, Zhou J, Sintim H O, Dayie T K 2014 Mol.Biosyst. 10 384
|
| [21] |
Kang M, Eichhorn C D, Feigon J 2014 Proc. Natl. Acad. Sci. USA 111 E663
|
| [22] |
Heroven A K, Nuss A M, Dersch P 2017 RNA Biol. 14 471
|
| [23] |
Wang H, Guan Z, Qiu J, Jia Y, Zeng C, Zhao Y 2020 RSC. Adv. 10 2004
|
| [24] |
Jiang L, Schaffitzel C, Bingel-Erlenmeyer R, Ban N, Korber P, Koning R I, de Geus Dd C, Plaisier J R, Abrahams J P 2009 J. Mol. Biol. 386 1357
|
| [25] |
Cate J H, Doudna J A 2000 Method. Enzymol. 317 169
|
| [26] |
Latham M P, Brown D J, McCallum S A, Prodi A 2005 Chembiochem 6 1492
|
| [27] |
Zhao Y, Wang J, Zeng C, Xiao Y 2018 Biophys. Rep. 4 123
|
| [28] |
Tang Y, Liu D, Wang Z, Wen T, Deng L 2017 BMC Bioinformatics 18 465
|
| [29] |
Su H, Liu M, Sun S, Peng Z, Yang J 2019 Bioinformatics 35 930
|
| [30] |
Duss O, Yulikov M, Jeschke G, Allain F H 2014 Nat. Commun. 5 3669
|
| [31] |
Duss O, Yulikov M, Allain F H T, Jeschke G 2015 Method. Enzymol. 558 279
|
| [32] |
Cheong H K, Hwang E, Lee C, Choi B S, Cheong C 2004 Nucleic Acids Res. 32 e84
|
| [33] |
Zhao Y, Huang Y, Zhou G, Wang Y, Man J, Xiao Y 2012 Sci. Rep. 2 734
|
| [34] |
Wang J, Xiao Y 2017 Current Protocols in Bioinformatics 57 5.9.1
|
| [35] |
Wang J, Wang J, Huang Y, Xiao Y 2019 Int. J. Mol. Sci. 20 4116
|
| [36] |
Gong Z, Zhao Y, Xiao Y 2010 J. Biomol. Struc. Dyn. 28 431
|
| [37] |
Zhao Y, Zhou G, Xiao Y 2011 J. Biomol. Struc. Dyn. 28 815
|
| [38] |
Sharma S, Ding F, Dokholyan N V 2008 Bioinformatics 24 1951
|
| [39] |
Krokhotin A, Houlihan K, Dokholyan N V 2015 Bioinformatics 31 2891
|
| [40] |
He J, Wang J, Tao H, Xiao Y, Huang S Y 2019 Nucleic Acids Res. 47 W35
|
| [41] |
Bao L, Zhang X, Jin L, Tan Z J 2015 Chin. Phys. B 25 018703
|
| [42] |
Siegfried N A, Busan S, Rice G M, Nelson J A, Weeks K M 2014 Nat. Methods 11 959
|
| [43] |
Harris M E, Christian E L 2009 Methods Enzymol. 468 127
|
| [44] |
Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A C, Munschauer M, Ulrich A, Wardle G S, Dewell S, Zavolan M, Tuschl T 2010 Cell 141 129
|
| [45] |
Zhao J, Ohsumi T K, Kung J T, Ogawa Y, Grau D J, Sarma K, Song J J, Kingston R E, Borowsky M, Lee J T 2010 Mol. Cell 40 939
|
| [46] |
Nilsen T W 2014 Cold Spring Harb. Protoc. 2014 683
|
| [47] |
Stork C, Zheng S 2018 Reporter Gene Assays Reporter Gene Assays New York Humana Press 1755 65 74
|
| [48] |
Shi Y Z, Wu Y Y, Wang F H, Tan Z J 2014 Chin. Phys. B 23 078701
|
| [49] |
Yang Y, Gu Q, Zhang B G, Shi Y Z, Shao Z G 2018 Chin. Phys. B 27 038701
|
| [50] |
Mueller F, Döring T, Erdemir T, Greuer B, Jünke N, Osswald M, Rinke-Appel J, Stade K, Thamm S, Brimacombe R 1995 Biochem. Cell Biol. 73 767
|
| [51] |
Massire C, Westhof E 1998 J. Mol. Graph. Model. 16 197
|
| [52] |
Jossinet F, Westhof E 2005 Bioinformatics 21 3320
|
| [53] |
Martinez H M, Maizel J V Jr, Shapiro B A 2008 J. Biomol. Struct. Dyn. 25 669
|
| [54] |
Jossinet F, Ludwig T E, Westhof E 2010 Bioinformatics 26 2057
|
| [55] |
Cao S, Chen S J 2005 RNA 11 1884
|
| [56] |
Parisien M, Major F 2008 Nature 452 51
|
| [57] |
Flores S C, Altman R B 2010 RNA 16 1769
|
| [58] |
Rother M, Rother K, Puton T, Bujnicki J M 2011 Nucleic Acids Res. 39 4007
|
| [59] |
Biesiada M, Purzycka K J, Szachniuk M, Blazewicz J, Adamiak R W 2016 RNA Structure Determination New York Humana Press 1490 199 215
|
| [60] |
Das R, Baker D 2007 Proc. Natl. Acad. Sci. USA 104 14664
|
| [61] |
Jonikas M A, Radmer R J, Laederach A, Das R, Pearlman S, Herschlag D, Altman R B 2009 RNA 15 189
|
| [62] |
Das R, Karanicolas J, Baker D 2010 Nat. Methods 7 291
|
| [63] |
Boniecki M J, Lach G, Dawson W K, Tomala K, Lukasz P, Soltysinski T, Rother K M, Bujnicki J M 2016 Nucleic Acids Res. 44 e63
|
| [64] |
Gutell R R, Power A, Hertz G Z, Putz E J, Stormo G D 1992 Nucleic Acids Res. 20 5785
|
| [65] |
Freyhult E, Moulton V, Gardner P 2005 Appl. Bioinformatics 4 53
|
| [66] |
Dunn S D, Wahl L M, Gloor G B 2008 Bioinformatics 24 333
|
| [67] |
Edgar R C 2004 Nucleic Acids Res. 32 1792
|
| [68] |
Chenna R, Sugawara H, Koike T, Lopez R, Gibson T J, Higgins D G, Thompson J D 2003 Nucleic Acids Res. 31 3497
|
| [69] |
Higgins D G, Sharp P M 1988 Gene 73 237
|
| [70] |
Katoh K, Kuma K, Toh H, Miyata T 2005 Nucleic Acids Res. 33 511
|
| [71] |
Edgar R C, Batzoglou S 2006 Curr. Opin. Struc. Biol. 16 368
|
| [72] |
Notredame C, Higgins D G, Heringa J 2000 J. Mol. Biol. 302 205
|
| [73] |
Lassmann T 2020 Bioinformatics 36 1928
|
| [74] |
Morcos F, Pagnani A, Lunt B, Bertolino A, Marks D S, Sander C, Zecchina R, Onuchic J N, Hwa T, Weigt M 2011 Proc. Natl. Acad. Sci. USA 108 E1293
|
| [75] |
De Leonardis E, Lutz B, Ratz S, Cocco S, Monasson R, Schug A, Weigt M 2015 Nucleic Acids Res. 43 10444
|
| [76] |
Weinreb C, Riesselman A J, Ingraham J B, Gross T, Sander C, Marks D S 2016 Cell 165 963
|
| [77] |
Weigt M, White R A, Szurmant H, Hoch J A, Hwa T 2009 PNAS. 106 67
|
| [78] |
Weinreb C, Riesselman A J, Ingraham J B, Gross T, Sander C, Marks D S 2016 Cell 165 963
|
| [79] |
Jian Y, Wang X, Qiu J, Wang H, Liu Z, Zhao Y, Zeng C 2019 BMC Bioinformatics 20 497
|
| [80] |
Hinton G E 2012 Neural Networks: Tricks of the Trade Berlin, Heidelberg Springer 7700 599 619
|
| [81] |
De Vries S J, Van Dijk M, Bonvin A M J J 2010 Nat. Protoc. 5 883
|
| [82] |
Trott O, Olson A J 2010 J. Comput. Chem. 31 445
|
| [83] |
Dominguez C, Boelens R, Bonvin A M 2003 J. Am. Chem. Soc. 125 1731
|
| [84] |
He J, Tao H, Huang S Y 2019 Bioinformatics 35 4994
|
| [85] |
Zeng P, Li J, Ma W, Cui Q 2015 Sci. Rep. 5 9179
|
| [86] |
Zeng P, Cui Q 2016 Sci. Rep. 6 19016
|
| [87] |
Wang K, Jian Y, Wang H, Zeng C, Zhao Y 2018 Bioinformatics 34 3131
|
| [88] |
Tuvshinjargal N, Lee W, Park B, Han K 2015 Comput. Meth. Prog. Bio. 120 3
|
| [89] |
Tuvshinjargal N, Lee W, Park B, Han K 2016 Biosystems 139 17
|
| [90] |
He X, Jun Wang, Wang J, Xiao Y 2020 Chin. Phys. B 29 078702
|
| [91] |
Griffithsjones S, Bateman A, Marshall M, Khanna A, Eddy S R 2003 Nucleic Acids Res. 31 439
|
| [92] |
Schulze-Gahmen U, Echeverria I, Stjepanovic G, Bai Y, Lu H, Schneidman-Duhovny D, Doudna J A, Zhou Q, Sali A, Hurley J H 2016 Elife 5 e15910
|
| [1] | Chengwei Deng(邓成伟), Yunxin Tang(唐蕴芯), Jian Zhang(张建), Wenfei Li(李文飞), Jun Wang(王骏), and Wei Wang(王炜). RNAGCN: RNA tertiary structure assessment with a graph convolutional network[J]. 中国物理B, 2022, 31(11): 118702-118702. |
| [2] | Wei-Bu Wang(王韦卜), Xing-Yuan Li(李兴元), and Ji-Guo Su(苏计国). Force-constant-decayed anisotropic network model: An improved method for predicting RNA flexibility[J]. 中国物理B, 2022, 31(6): 68704-068704. |
| [3] | Yan-Hui Li(李彦慧), Zhen-Sheng Zhong(钟振声), and Jie Ma(马杰). Single-molecule mechanical folding and unfolding kinetics of armless mitochondrial tRNAArg from Romanomermis culicivorax[J]. 中国物理B, 2021, 30(10): 108203-108203. |
| [4] | Wen-Jing Wang(王文静) and Ji-Guo Su(苏计国). Enhancements of the Gaussian network model in describing nucleotide residue fluctuations for RNA[J]. 中国物理B, 2021, 30(5): 58701-058701. |
| [5] | . [J]. 中国物理B, 2021, 30(2): 28705-0. |
| [6] | Bin Huang(黄斌), Yuanyang Du(杜渊洋), Shuai Zhang(张帅), Wenfei Li(李文飞), Jun Wang(王骏), Jian Zhang(张建). [J]. 中国物理B, 2020, 29(10): 108704-. |
| [7] | Zhi-Chao Liu(刘志超), Qin Liu(刘琴), Chan-You Chen(陈禅友), Chen Zeng(曾辰), Peng Ran(冉鹏), Yun-Jie Zhao(赵蕴杰), Lei Pan(潘磊). [J]. 中国物理B, 2020, 29(10): 108709-. |
| [8] | Sha Gong(龚沙), Taigang Liu(刘太刚), Yanli Wang(王晏莉), Wenbing Zhang(张文炳). [J]. 中国物理B, 2020, 29(10): 108703-. |
| [9] | 何小玲, 王军, 王剑, 肖奕. Improving RNA secondary structure prediction using direct coupling analysis[J]. 中国物理B, 2020, 29(7): 78702-078702. |
| [10] | 杨毅, 辜琦, 张本龚, 时亚洲, 邵志刚. A novel knowledge-based potential for RNA 3D structure evaluation[J]. 中国物理B, 2018, 27(3): 38701-038701. |
| [11] | 熊贵, 席昆, 张曦, 谭志杰. To what extent of ion neutralization can multivalent ion distributions around RNA-like macroions be described by Poisson-Boltzmann theory?[J]. 中国物理B, 2018, 27(1): 18203-018203. |
| [12] | 王宇杰, 王珍, 王晏莉, 张文炳. Molecular dynamic simulation of the thermodynamic and kinetic properties of nucleotide base pair[J]. 中国物理B, 2017, 26(12): 128705-128705. |
| [13] | 常晓雪, 徐留芳, 史华林. Theoretical studies on sRNA-mediated regulation in bacteria[J]. 中国物理B, 2015, 24(12): 128703-128703. |
| [14] | 时亚洲, 吴园燕, 王凤华, 谭志杰. RNA structure prediction:Progress and perspective[J]. 中国物理B, 2014, 23(7): 78701-078701. |
|
||