中国物理B ›› 2020, Vol. 29 ›› Issue (10): 108702-.doi: 10.1088/1674-1056/abb225
Xiang Li(李翔)1,2, Hong Qi(祁宏)3, Xiao-Cui Zhang(张晓翠)1, Fei Xu(徐飞)1, Zhi-Yong Yin(尹智勇)1, Shi-Yang Huang(黄世阳)4, Zhao-Shou Wang(王兆守)4,†( ), Jian-Wei Shuai(帅建伟)1,2,5,‡(
), Jian-Wei Shuai(帅建伟)1,2,5,‡( )
)
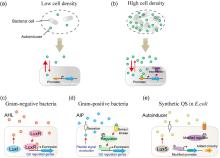
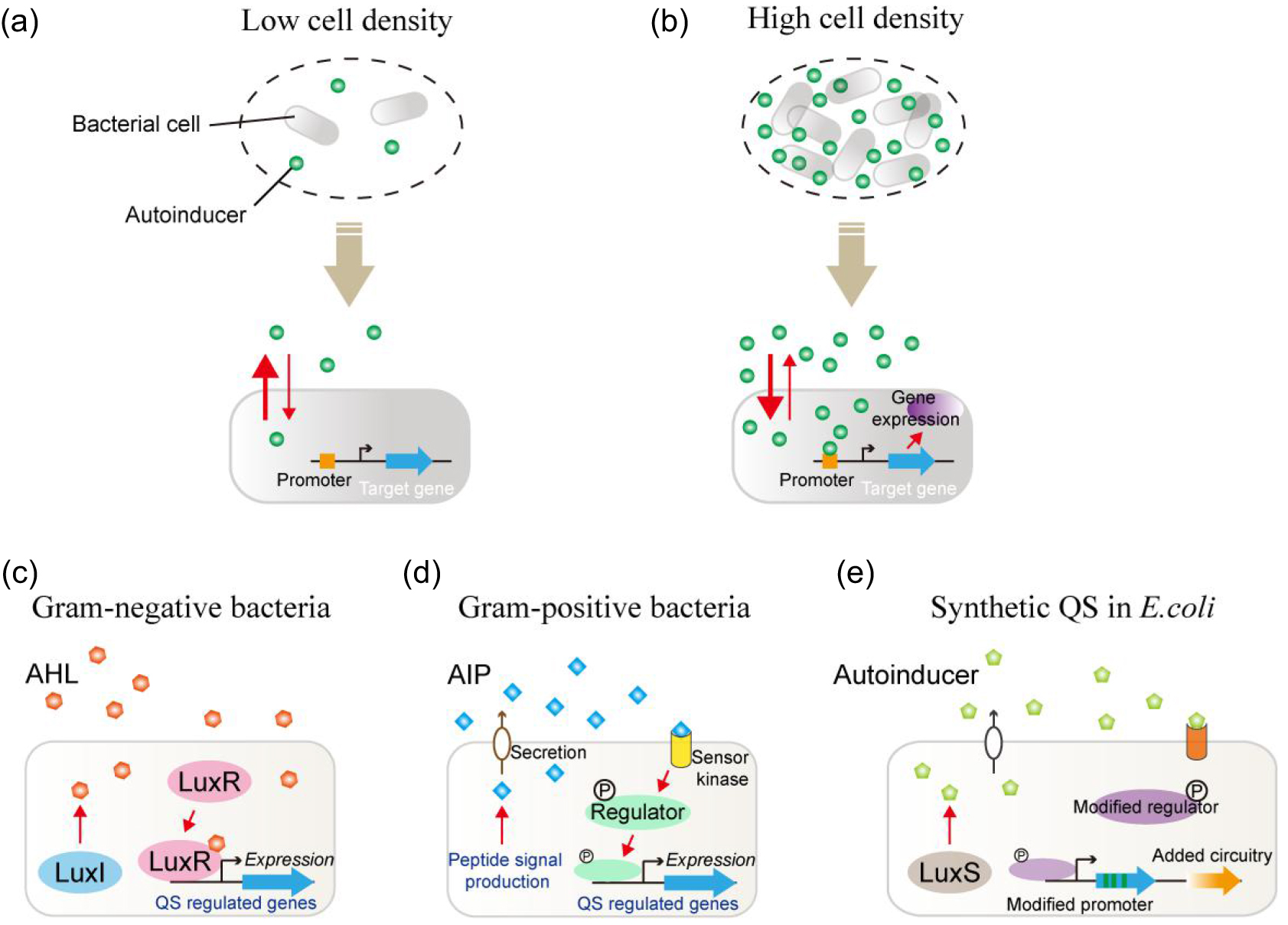
Quantitative modeling of bacterial quorum sensing dynamics in time and space
Xiang Li(李翔)1,2, Hong Qi(祁宏)3, Xiao-Cui Zhang(张晓翠)1, Fei Xu(徐飞)1, Zhi-Yong Yin(尹智勇)1, Shi-Yang Huang(黄世阳)4, Zhao-Shou Wang(王兆守)4,†, and Jian-Wei Shuai(帅建伟)1,2,5,‡
- 1
Department of Physics, College of Physical Science and Technology, Xiamen University , Xiamen 361005,China
2State Key Laboratory of Cellular Stress Biology, Innovation Center for Cell Signaling Network, Xiamen University , Xiamen 361102,China
3Complex Systems Research Center, Shanxi University , Taiyuan 030006,China
4Institute of Biochemical Engineering, Department of Chemical and Biochemical Engineering, College of Chemistry and Chemical Engineering, Xiamen University , Xiamen 361005,China
5National Institute for Data Science in Health and Medicine, Xiamen University , Xiamen 361102,China