†Corresponding author. E-mail: lupingz@uow.edu.au
‡Corresponding author. E-mail: xbning@nju.edu.cn
*Project supported by the National Natural Science Foundation of China (Grant No. 61271079), the Vice Chancellor Research Grant in University of Wollongong, and the Priority Academic Program Development of Jiangsu Higher Education Institutions, China.
Applications of multifractal analysis to white matter structure changes on magnetic resonance imaging (MRI) have recently received increasing attentions. Although some progresses have been made, there is no evident study on applying multifractal analysis to evaluate the white matter structural changes on MRI for Alzheimer’s disease (AD) research. In this paper, to explore multifractal analysis of white matter structural changes on 3D MRI volumes between normal aging and early AD, we not only extend the traditional box-counting multifractal analysis (BCMA) into the 3D case, but also propose a modified integer ratio based BCMA (IRBCMA) algorithm to compensate for the rigid division rule in BCMA. We verify multifractal characteristics in 3D white matter MRI volumes. In addition to the previously well studied multifractal feature, Δ α, we also demonstrated Δ f as an alternative and effective multifractal feature to distinguish NC from AD subjects. Both Δ α and Δ f are found to have strong positive correlation with the clinical MMSE scores with statistical significance. Moreover, the proposed IRBCMA can be an alternative and more accurate algorithm for 3D volume analysis. Our findings highlight the potential usefulness of multifractal analysis, which may contribute to clarify some aspects of the etiology of AD through detection of structural changes in white matter.
Alzheimer’ s disease (AD) is affecting millions of people, especially the elderly around the world.[1] Histopathologic and magnetic resonance imaging (MRI) studies have indicated that white matter changes, such as loss of myelin and axons and selective changes in lipid components, can be deemed as one contributing factor to the pathology of AD.[2, 3] Since there is still no absolute cure for AD, it could be helpful and significant for early AD detection by exploring the white matter structure changes on MRI.
Conventional methods, such as neuropathological assessment, [4] cortical thickness, [5] voxel-based morphometry, [6] have identified white matter abnormality in AD subjects. However, these methods ignore the intrinsic fluctuation pattern in the white matter signal. Considering the heterogeneities caused by the loss of white matter tissues, it might be difficult to be evaluated by visual inspection or the conventional texture-based analysis.[7]
Fractal, which can characterize a self-similar structure, has been widely used to describe geometrical properties of morphological complexity and also been observed in white matter.[8] In contrast to simple fractals, the concept of multifractal holds that different regions of an object have different local fractal properties, [9] which can give a more comprehensive description for the object. Consequently, multifractal analysis, which can provide a precise quantitative description of a broad range of heterogeneous phenomena by measuring the intrinsic fluctuation pattern, has recently been given considerable attentions.[7, 10– 14] Takahashi and his colleagues have successfully employed the standard partition function multifractal formalism to quantitatively evaluate the white matter structural changes in normal aging, [10] the early-stage atherosclerosis, [7] and schizophrenia.[11] However, to the best of our knowledge, no evident study has been conducted on applying multifractal analysis to evaluate the white matter structural changes on MRI for AD research.
Moreover, there are two main limitations in the current studies.[7, 10, 11] Firstly, the multifractal analysis used in the existing approaches was only restricted on the single or consecutive 2D slice images, while 3D information is very important in medical image analysis referring to volumetric objects.[15] For example, Chen et al.[16] studied breast MRI data by 3D gray-level co-occurrence matrix, and demonstrated that the classification performance of volumetric features was significantly better than those based on 2D analysis. Hence, to achieve a precise description of the structural complexity of white matter, three-dimensional multifractal analysis might be preferred. Secondly, when calculating the multifractal spectrum in Refs. [7], [10], and [11], only the box sizes of 2n were considered, which cannot handle the fractal dimensions with arbitrary box size and might miss useful information.
In this paper, we aim to investigate: (i) whether there exist multifractal characteristics in 3D MRI volume; (ii) whether the multifractal analysis can be used on 3D MRI volume to distinguish normal aging from the early AD. To accomplish these objectives, we carry out a secondary analysis on the high-resolution MRI data from the publicly available dataset Alzheimer’ s Disease Neuroimaging Initiative (ADNI). Besides, in addition to extend the traditional 2D box-counting multifractal analysis (BCMA) into the 3D case, we also proposed a modified integer ratio based 3D BCMA (IRBCMA) algorithm, which can compensate for the rigid division rule (2n) in BCMA.
The rest of this article is organized as follows. Section 2 introduces the 3D extension of traditional BCMA and proposes a modified IRBCMA algorithm for more accurate calculation. This is followed by multifractal feature extraction. In Section 3, experimental verifications of the proposed methods on ADNI dataset were conducted. The information for participants and data preprocessing is firstly provided in Section 3.1. Afterwards, multifractal characteristics are verified in Section 3.2. In Section 3.3, the viability of multifractal features for AD detection was investigated and discussed. Section 4 concludes this article.
The essence of multifractal analysis is to constitute the multifractal spectrum (MFS), which can describe the complexity of the object. Box-counting method is the simplest and effective way for multifractal analysis and has been successfully applied to the research on white matter structural changes.[7, 10, 11] In this study, the traditional 2D BCMA[7] was extended into 3D analysis. More importantly, to circumvent the aforementioned restricted division rule in BCMA, we, inspired by Ref. [17], proposed the integer ratio based 3D BCMA (IRBCMA) to measure the complex dimensions of volumes.
The multifractal analysis modified by Chhabra et al.[18] is used for 3D BCMA and a typical MFS is shown in Fig. 1(a). Briefly, after dividing the 3D object into cubes with sizes δ × δ × δ , we assign a probability to each cube as 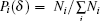

where n is the number of the segmented cubes, and q can vary between − ∞ to + ∞ theoretically. Next, from the perspective of entropy, the singularity strength α (q) is given by
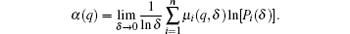
Meanwhile, the fractal dimension f(q) is given by
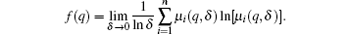
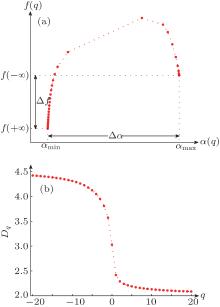
The plot of f(q) versus α (q) is called MFS, which is generally a hook-shaped curve, as illustrated in Fig. 1(a). In traditional BCMA, the “ power of 2” cube size (i.e., δ = 2n) is needed to divide the object for 3D MFS calculation in different scales.[7] In this study, δ = 1, 2, 4, 8, 16 is used.
It should be noted that the value of q cannot be infinite in practice. In fact, if α (q) and f(q) tend to be constant, the maximum | q| can be deemed as the approximation limit of the endpoints. Following this idea, − 20 ≤ q ≤ 20 is used in this study. When q = − 20, the α (q) value is denoted as α max, while the f(q) value is denoted as f(− ∞ ). Accordingly, the α min and f(+ ∞ ) values are defined by α (q) and f(q) at q = 20, respectively. The positions of α max, f(− ∞ ), α min and f(+ ∞ ) are illustrated in Fig. 1(a).
In the traditional BCMA, only the divisions with δ = 2n are considered, which may omit some useful information beyond the restricted division size. Hence, we propose the 3D IRBCMA for calculation under arbitrary sizes.
Given a 3D object I with M × N × K voxels, for any ratio r(r ≥ 2, r ∈ Z+ ), we superpose a grid of blocks with m × n × k voxels, where m = ⌊ M/r⌋ , n = ⌊ N/r ⌋ , and k = ⌊ K/r ⌋ . Based on whether the values of M, N, and K could be exactly divided by r, there are eight situations to deal with.
Case 1M = mr, N = nr, and K = kr. The object is evenly partitioned into r × r × r blocks of m × n × k voxels.
Case 2M = mr, N = nr, and K > kr. The object is partitioned into r × r × (r + 1) blocks. Among these blocks, there are r × r × r blocks of m × n × k voxels and r × r × 1 blocks of m × n × (K − kr) voxels.
Case 3M = mr, N > nr, and K = kr. The object is partitioned into r × (r + 1) × r blocks. Among these blocks, there are r × r × r blocks of m × n × k voxels and r × 1 × r blocks of m × (N − nr) × k voxels.
Case 4M > mr, N = nr, and K = kr. The object is partitioned into (r + 1) × r × r blocks. Among these blocks, there are r × r × r blocks of m × n × k voxels and 1 × r × r blocks of (M − mr) × n × k voxels.
Case 5M = mr, N > nr, and K > kr. The object is partitioned into r × (r + 1) × (r + 1) blocks. Among these blocks, there are r × r × r blocks of m × n × k voxels, r × r × 1 blocks of m × n × (K − kr) voxels, r × 1 × r blocks of m × (N − nr) × k voxels, and r × 1 × 1 blocks of m × (N − nr) × (K − kr) voxels.
Case 6M > mr, N = nr, and K > kr. The object is partitioned into (r + 1) × r × (r + 1) blocks. Among these blocks, there are r × r × r blocks of m × n × k voxels, r × r × 1 blocks of m × n × (K − kr) voxels, 1 × r × r blocks of (M − mr) × n × k voxels, and 1 × r × 1 blocks of (M − mr)× n × (K − kr) voxels.
Case 7M > mr, N > nr, and K = kr. The object is partitioned into (r + 1) × (r + 1) × r blocks. Among these blocks, there are r × r × r blocks of m × n × k voxels, r × 1 × r blocks of m × (N − nr) × k voxels, 1 × r × r blocks of (M − mr) × n × k voxels, and 1 × 1 × r blocks of (M − mr)× (N − nr) × k voxels.
Case 8M > mr, N > nr, and K > kr. The object is partitioned into (r + 1) × (r + 1) × (r + 1) blocks. Among these blocks, there are r × r × r blocks of m × n × k voxels, r × r × 1 blocks of m × n × (K − kr) voxels, r × 1 × r blocks of m × (N − nr) × k voxels, 1 × r × r blocks of (M − mr) × n × k voxels, r × 1 × 1 blocks of m × (N − nr) × (K − kr) voxels, 1 × r × 1 blocks of (M − mr) × n × (K − kr) voxels, 1 × 1 × r blocks of (M − mr) × (N − nr) × k voxels, and 1 × 1 × 1 blocks of (M − mr) × (N − nr) × (K − kr) voxels.
After partitioning the object, for a certain ratio r, we define the intensity value T(i, j, k) in the (i, j, k)-th block. In order to contain the volume tightly and accurately, we allow the intensity value to be real, rather than integer. In addition, instead of using the difference of the maximum and minimum values in one block like Eq. (8) in Ref. [17], we take the sum of values as the measure, which can reflect more properties in the block. Thus, the intensity value in the (i, j, k)-th block is calculated as

where V(i, j, k) is the volume of the (i, j, k)-th block. The value of V(i, j, k) is equal to m × n × k for most blocks, but smaller for the remainder blocks along the object’ s borders. Next, similar to the BCMA in Section 2.1, the probability in the (i, j, k)-th block is measured by
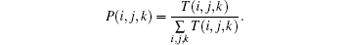
The normalized q-th moment of the probability density is
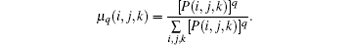
Afterwards, as derived in Ref. [18], the singularity strength is then given by
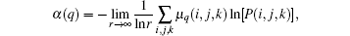
and the fractal dimension can be calculated by
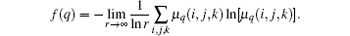
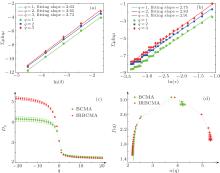
Generally, two strategies are commonly used for verifying multifractal characteristics in the box-counting method. In Ref. [18], different slopes from linearly fitting of ∑ μ lnμ with respect to lnL for different q values (usually q = 1, 2, 3) has been used as a simple and effective way to detect multifractal characteristics. In the BCMA case (see Fig. 3(a)), it refers to ∑ μ i(q, δ ) ln μ i(q, δ ) with respect to ln δ , while ∑ μ q(i, j, k) ln μ q (i, j, k) with respect to ln r is referred in the IRBCMA case (see Fig. 3(b)). Moreover, the “ generalized dimensions” , Dq, which correspond to scaling exponents for the q-th moments of the measure, can be achieved by
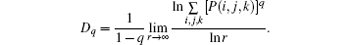
For an object with significant multifractal characteristics, the plot of Dq versus q is generally sigmoidal and decreasing.[19] A typical illustration is shown in Fig. 1(b).
According to the principle described in Ref. [20], the number of pixels covered by a particular grid block should be no less than the maximum number of the boxes in that block. Similar to this idea, the box size in the 3D IRBCMA method should meet the criteria of m × n × k = M/r × N/r × K/r ≥ r, which means 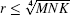
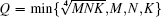
For both BCMA and IRBCMA, two canonical features, Δ α and Δ f, are selected to capture the multifractal properties of white matter structure, which are shown in Fig. 1(a) as illustration.
The feature Δ α = α max − α min represents the width of the MFS, which can detect the degree of multifractality of the object. In the multifractal formalism, [9, 18] the distribution probability in the m-th box can be measured by Pm(r) ∝ (1/r)α m, where r (usually 0 < 1/r < 1) is the integer ratio forming the block size and α m is the singularity strength of the probability subset. Obviously, Pm(r) increases with decreasing α m at a fixed r. The value α max is related to the minimum probability measure via Pmin ∝ (1/r)α max, which corresponds to the smallest intensity values at scale 1/r. In contrast, α min, measured via Pmax ∝ Lα min, corresponds to the largest intensity values at scale 1/r. The Δ α can thus be used to describe the range of the probability measures with Pmax/Pmin ∝ rΔ α . The greater the Δ α value is, the wider the intensity distribution, the richer the structure, and the stronger the degree of multifractality in the object.
The feature Δ f = f(+ ∞ ) − f(− ∞ ) represents the difference of fractal dimension. In the formalism of Refs. [9] and [18], f(+ ∞ ) reflects the fractal dimension of the subset of the maximum probability with NPmax = Nα min ∝ (1/r)− f(+ ∞ ), indicating the number of the largest intensity values, while f(− ∞ ) reflects the number of the smallest intensity values. Hence, the Δ f value can describe the ratio between the largest and the smallest intensity regions of the object: NPmax/NPmin ∝ (1/L)Δ f.
MRI data used in this study were downloaded from the ADNI database. The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations as a $ 60 million, 5-year public private partnership. The primary goal of ADNI has been to test whether the combination of serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The principal investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. The ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations. The study subjects were recruited from over 50 sites across the US and Canada. They gave the written informed consent at the time of enrolment for imaging and genetic sample collection and completed the questionnaires approved by the Institutional Review Board (IRB) of each participating site.
Thirty-eight normal elderly controls (NC) (mean± SD: (77.2± 6.5) years) and twenty-five patients with early AD (mean± SD: (73.7± 7.6) years) were included. Among the subjects, twenty-eight NC and eleven early AD subjects were recorded with mini-mental state examination (MMSE) scores, which are extensively used to measure cognitive impairment and the dementia degree. The lower MMSE score is related to more severe cognitive impairment or dementia of the subject.[21] The sagittal 3D T1-weighted magnetization prepared rapid gradient echo (MPRAGE) images with a resolution of 1 × 1 × 1.2 mm3 were acquired (TR = 6.8 ms; TE = 3.16 ms; flip angle is 9° ; thickness is 1.2 mm; matrix is 256 × 256).
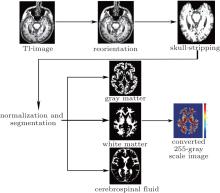
The standard preprocessing is shown in Fig. 2, which is performed using SPM8. The steps include manual reorientation, skull-stripping, spatial normalization to the MNI space, and segmentation into gray matter, white matter, and cerebrospinal fluid by employing diffeomorphic anatomical registration using the exponentiated liealgebra (DARTEL) method.[22] Considering that the multifractal analysis evaluates the fluctuation of the objects, the MRI signal intensities were normalized and converted into 255-gray scale values to exclude the adverse effect of possible different intensity ranges among subjects. After preprocessing, the automatically segmented white matter was used for the further multifractal analysis.
In this study, based on the preprocessed 3D MRI volumes of white matter, we firstly verified their multifractal characteristics by plotting the three fitted lines of ∑ μ lnμ with respect to lnL for q = 1, 2, 3 and the curves of Dq versus q for both BCMA and IRBCMA. The typical illustrations are shown in Fig. 3(a)– 3(c). Indeed for all subjects, the slopes of the three fitted lines for q = 1, 2, 3 are different and the Dq versus q curves tend to be sigmoidal and decreasing, which demonstrate that the 3D white matter volumes can be characterized by multifractals.
In our experiment, the averaged results from different divisions are used for more accurate multifractal results. The mean ± standard error for both BCMA and IRBCMA on MFS are plotted in Fig. 3(d). Multifractal features, Δ α and Δ f, were extracted based on the MFS. For both BCMA and IRBCMA, the statistic test results for both NC and AD groups are shown in Table 1. It clearly shows significant differences between NC and AD groups (p < 0.05) for both BCMA and IRBCMA, which suggests that Δ α and Δ f from both methods can effectively distinguish the NC and AD groups on 3D white matter MRI structure.
| Table 1. Results of multifractal features for NC and AD groups. |
In the previous studies, [7, 10, 11] only Δ α was reported to be a suitable multifractal feature of heterogeneity and used for exploring the white matter structural changes on MRI. As the measure for detecting multifractal degree, Δ α has been demonstrated to be effective in our study again. From Table 1, the larger Δ α value in NC group indicates that the NC group has wider intensity distributions and stronger multifractal structures than the AD counterpart, which is in line with the previous report of the smaller Δ α observed in patients.[11] In addition to Δ α , the feature Δ f was also found to be sensitive in distinguishing NC and AD groups, which may suggest Δ f can be an alternative feature on such study. The Δ f value can describe the ratio between the largest and the smallest intensity regions of the white matter volume. In Table 1, Δ f < 0 is found for both NC and AD groups, which results from the small percentage of the white matter volume in the whole brain and hence the smaller number of regions with large intensity. Moreover, the Δ f values in NC group are larger than those in AD group, which may imply that in the brains, the proportion of the white matter volume in NC is larger than that in AD group. In Ref. [6], Migliaccio et al. employed the voxel-based morphometric analysis on NC and AD groups, and they found white matter atrophy in AD group. Our finding is in line with this report.
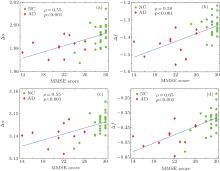
On the other hand, Pearson’ s correlation coefficient (ρ ) was used to test the association between the clinical MMSE scores and the multifractal features, as shown in Fig. 4. Strong positive correlations with statistical significance were found for both Δ α and Δ f with MMSE scores (ρ > 0.5 and p < 0.05). It is clearly suggested that the extracted multifractal features, Δ α and Δ f, are closely correlated with the clinical MMSE scores. Lower MMSE scores in AD subjects correspond to smaller Δ α and Δ f values, indicating the decline of complexity of white matter structure in the AD group.
It should be noted that both BCMA and IRBCMA were employed in this study. The traditional BCMA in the 2D case has been successfully applied for white matter structural changes on MRI.[7, 10, 11] In this study, we extended it into the 3D case and obtained satisfactory results. However, BCMA is restricted to be calculated only on box size with 2n, which may neglect some useful information. Hence, we proposed the modified IRBCMA algorithm, which can relax the division to arbitrary box size. Our work was underpinned by the intuition that, the IRBCMA method can count boxes more accurately by capturing comprehensive information from arbitrary division. This is supported by the finding of the stronger correlation between Δ f and MMSE (ρ = 0.65, p < 0.05) by the IRBCMA compared with that obtained by BCMA (ρ = 0.59, p < 0.05), as shown in Figs. 4(b) and 4(d). Therefore, it may suggest that IRBCMA can be an alternative and more accurate algorithm for 3D volume analysis.
In this study, we explored multifractal analysis of white matter structural changes on 3D MRI volumes between normal aging and early AD. Based on the MFS, the Δ α and Δ f, as two canonical multifractal features, were extracted and demonstrated as the effective features to distinguish NC from AD subjects. We highlight that Δ f is found to be an alternative feature for multifractal analysis on MRI data, while in the previous studies only Δ α was used. Moreover, in addition to extend the traditional BCMA into the 3D case, we also proposed a modified IRBCMA algorithm, which can compensate for the rigid division rule (2n) in BCMA. Superior performances from both BCMA and IRBCMA have been obtained in this study, which suggests that IRBCMA can be an alternative algorithm for 3D volume analysis. It should be noted that, possibly more accurate and effective Δ f values can be obtained by IRBCMA, as indicated by the stronger correlation between Δ f and MMSE. Both Δ α and Δ f with respect to the clinical MMSE scores are found to have strong positive correlations with statistical significance, which ensures the potential physiological meaning for Δ α and Δ f. Thus, investigations, such as the present study, may be important steps in direction towards finding practically effective marker for AD detection. Our findings highlight the potential usefulness of multifractal analysis, which may contribute to clarifying some aspects of the etiology of AD through detection of structural changes in white matter.
| 1 |
|
| 2 |
|
| 3 |
|
| 4 |
|
| 5 |
|
| 6 |
|
| 7 |
|
| 8 |
|
| 9 |
|
| 10 |
|
| 11 |
|
| 12 |
|
| 13 |
|
| 14 |
|
| 15 |
|
| 16 |
|
| 17 |
|
| 18 |
|
| 19 |
|
| 20 |
|
| 21 |
|
| 22 |
|