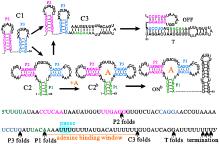
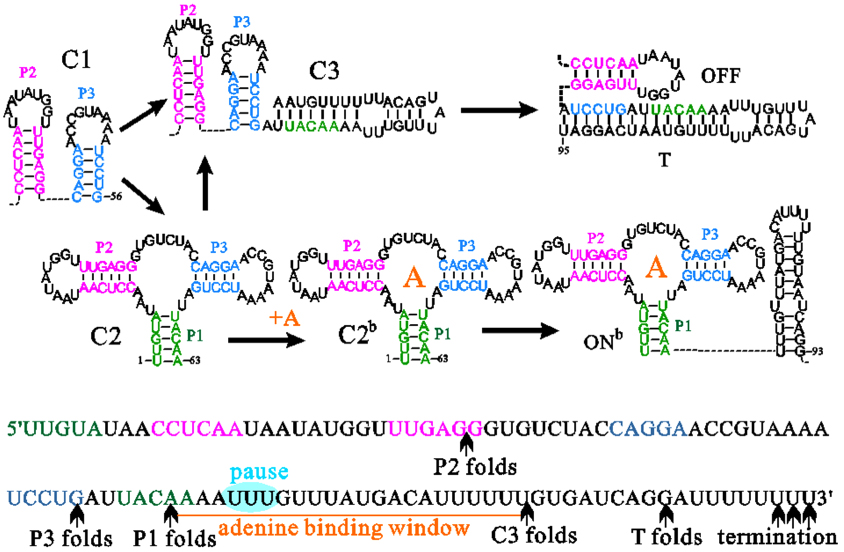
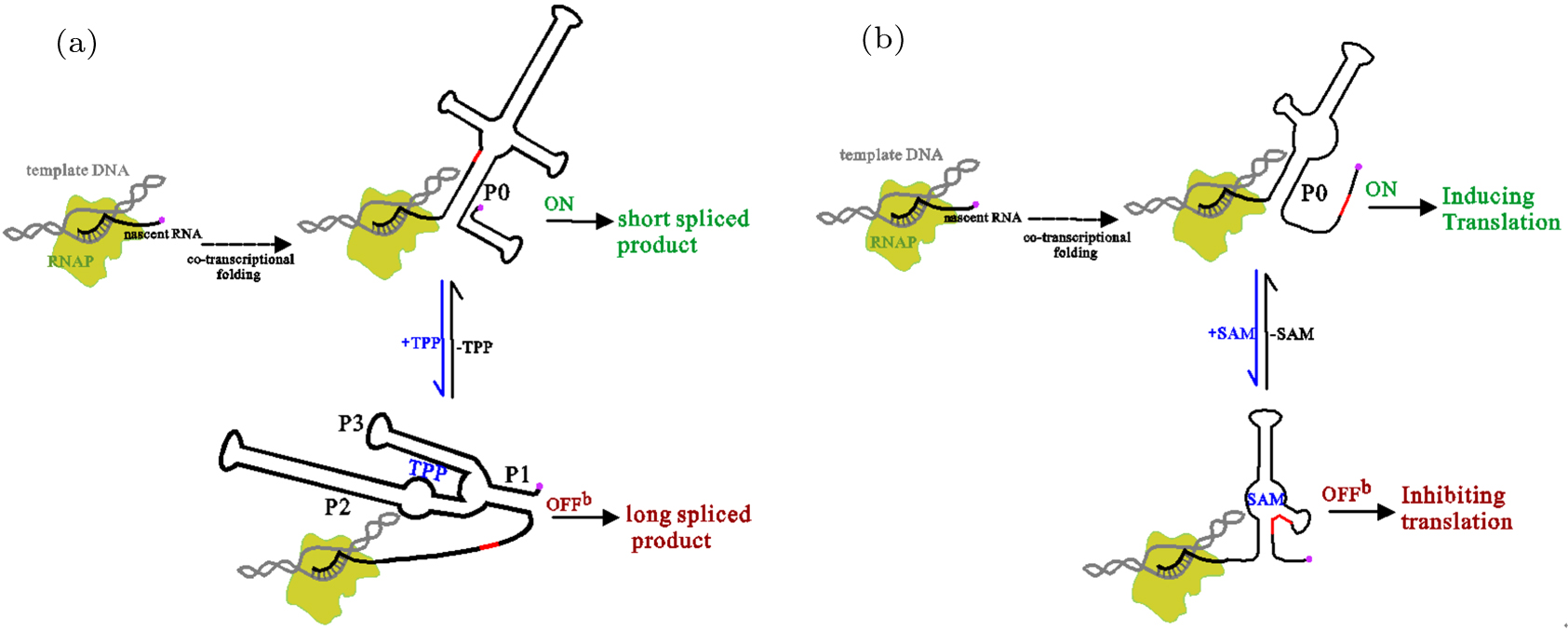
中国物理B ›› 2020, Vol. 29 ›› Issue (10): 108703-.doi: 10.1088/1674-1056/abab84
Sha Gong(龚沙)1, Taigang Liu(刘太刚)2, Yanli Wang(王晏莉)2, Wenbing Zhang(张文炳)2,†( )
)
-
收稿日期:2020-06-29修回日期:2020-07-30接受日期:2020-08-01出版日期:2020-10-05发布日期:2020-10-05 -
通讯作者:Wenbing Zhang(张文炳)
The theory of helix-based RNA folding kinetics and its application
Sha Gong(龚沙)1, Taigang Liu(刘太刚)2, Yanli Wang(王晏莉)2, and Wenbing Zhang(张文炳)2,†
- 1
Hubei Key Laboratory of Economic Forest Germplasm Improvement and Resources Comprehensive Utilization, Hubei Collaborative Innovation Center for the Characteristic Resources Exploitation of Dabie Mountains, Huanggang Normal University , Huanggang 438000,China
2Department of Physics, Wuhan University , Wuhan 430072,China
-
Received:2020-06-29Revised:2020-07-30Accepted:2020-08-01Online:2020-10-05Published:2020-10-05 -
Contact:†Corresponding author. E-mail:wbzhang@whu.edu.cn -
About author:†Corresponding author. E-mail: wbzhang@whu.edu.cn* Project supported by the Science Fund from the Key Laboratory of Hubei Province, China (Grant No. 201932003) and the National Natural Science Foundation of China (Grant Nos. 1157324 and 31600592).
中图分类号: (RNA)
- 87.14.gn
引用本文
Sha Gong(龚沙), Taigang Liu(刘太刚), Yanli Wang(王晏莉), Wenbing Zhang(张文炳). [J]. 中国物理B, 2020, 29(10): 108703-.
Sha Gong(龚沙), Taigang Liu(刘太刚), Yanli Wang(王晏莉), and Wenbing Zhang(张文炳)†. The theory of helix-based RNA folding kinetics and its application[J]. Chin. Phys. B, 2020, 29(10): 108703-.
| [1] |
Zhuang X 2000 Science 288 2048 DOI: 10.1126/science.288.5473.2048
|
| [2] |
Strulson C A, Molden R C, Keating C D, Bevilacqua P C 2012 Nat. Chem. 4 941 DOI: 10.1038/nchem.1466
|
| [3] |
Das R, Karanicolas J, Baker D 2010 Nat. Methods 7 291 DOI: 10.1038/nmeth.1433
|
| [4] |
Förster U, Weigand J E, Trojanowski P, Suess B, Wachtveitl J 2012 Nucleic Acids Res. 40 1807 DOI: 10.1093/nar/gkr835
|
| [5] |
Gong B, Klein D 2011 J. Am. Chem. Soc. 133 14188 DOI: 10.1021/ja205185g
|
| [6] |
Marraffini L A, Sontheimer E J 2010 Nat. Rev. Genet. 11 181 DOI: 10.1038/ncomms8552
|
| [7] |
Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A 2000 Cell 102 615 DOI: 10.1016/S0092-8674(00)00084-2
|
| [8] |
Nissen P, Hansen J, Ban N, Moore P B, Steitz T A 2000 Science 289 920 DOI: 10.1126/science.289.5481.920
|
| [9] |
Ahmad S, Muthukumar S, Kuncha S K, Routh S B, Yerabham A S K, Hussain T, Kamarthapu V, Kruparani S P, Sankaranarayanan R 2015 Nat. Commun. 6 1 DOI: 10.1038/ncomms8552
|
| [10] |
Zhong G, Wang H, He W, Li Y, Mou H, Tickner Z J, Tran M H, Ou T, Yin Y, Diao H, Farzan M 2020 Nat. Biotechnol. 38 169 DOI: 10.1038/s41587-019-0357-y
|
| [11] |
Wimberly B T, Brodersen D E, Clemons W M, Morgan-Warren R J, Carter A P, Vonrhein C, Hartsch T, Ramakrishnan V 2000 Nature 407 327 DOI: 10.1038/35030006
|
| [12] |
Herschlag D 1995 J. Biol. Chem. 270 20871 DOI: 10.1074/jbc.270.36.20871
|
| [13] |
Geis M, Flamm C, Wolfinger M T, Tanzer A, Hofacker I L, Middendorf M, Mandl C, Stadler P F, Thurner C 2008 J. Mol. Biol. 379 160 DOI: 10.1016/j.jmb.2008.02.064
|
| [14] |
Thirumalai D, Hyeon C 2005 Biochemistry 44 4957 DOI: 10.1021/bi047314+
|
| [15] |
Frieda K L, Block S M 2012 Science 338 397 DOI: 10.1126/science.1225722
|
| [16] |
Gong S, Wang Y J, Zhang W B 2015 J. Chem. Phys. 143 045103 DOI: 10.1063/1.4927390
|
| [17] |
Poot R A, Tsareva N V, Boni I V, van Duin J 1997 Proc. Natl. Acad. Sci. 94 10110 DOI: 10.1073/pnas.94.19.10110
|
| [18] |
Gerdes K, Wagner E G H 2007 Curr. Opin. Microbiol. 10 117 DOI: 10.1016/j.mib.2007.03.003
|
| [19] |
Ren A, Rajashankar K R, Patel D J 2012 Nature 486 85 DOI: 10.1038/nature11152
|
| [20] |
Zemora G, Waldsich C 2010 RNA Biol. 7 634 DOI: 10.4161/rna.7.6.13554
|
| [21] |
DebRoy S, Gebbie M, Ramesh A, Goodson J R, Cruz M R, van Hoof A, Winkler W C, Garsin D A 2014 Science 345 937 DOI: 10.1126/science.1255091
|
| [22] |
Lemay J F, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Massé E, Lafontaine D A 2011 PLoS Genet. 7 e1001278 DOI: 10.1371/journal.pgen.1001278
|
| [23] |
Schroeder R, Grossberger R, Pichler A, Waldsich C 2002 Curr. Opin. Struct. Biol. 12 296 DOI: 10.1016/S0959-440X(02)00325-1
|
| [24] |
Wong T N, Pan T 2009 Methods Enzymol. 468 167 DOI: 10.1016/S0076-6879(09)68009-5
|
| [25] |
Lubkowska L, Maharjan A S, Komissarova N 2011 J. Biol. Chem. 286 31576 DOI: 10.1074/jbc.M111.249359
|
| [26] |
Boyle J, Robillard G T, Kim S H 1980 J. Mol. Biol. 139 601 DOI: 10.1016/0022-2836(80)90051-0
|
| [27] |
Nussinov R, Tinoco I 1981 J. Mol. Biol. 151 519 DOI: 10.1016/0022-2836(81)90008-5
|
| [28] |
Zhang L, Bao P, Leibowitz M J, Zhang Y 2009 RNA 15 1986 DOI: 10.1261/rna.1638609
|
| [29] |
Wong T N, Sosnick T R, Pan T 2007 Proc. Natl. Acad. Sci. 104 17995 DOI: 10.1073/pnas.0705038104
|
| [30] |
Pan T, Artsimovitch I, Fang X W, Landick R, Sosnick T R 1999 Proc. Natl. Acad. Sci. 96 9545 DOI: 10.1073/pnas.96.17.9545
|
| [31] |
Heilman-Miller S L, Woodson S A 2003 RNA 9 722 DOI: 10.1261/rna.5200903
|
| [32] |
Cech T R 1990 Ann. Rev. Biochem. 59 543 DOI: 10.1146/annurev.bi.59.070190.002551
|
| [33] |
Michel F 1995 Ann. Rev. Biochem. 64 435 DOI: 10.1146/annurev.bi.64.070195.002251
|
| [34] |
Lutz B, Faber M, Verma A, Klumpp S, Schug A 2014 Nucleic Acids Res. 42 2687 DOI: 10.1093/nar/gkt1213
|
| [35] |
Sauerwine B, Widom M 2011 Phys. Rev. E 84 061912 DOI: 10.1103/PhysRevE.84.061912
|
| [36] |
Faber M, Klumpp S 2013 Phys. Rev. E 88 052701 DOI: 10.1103/PhysRevE.88.052701
|
| [37] |
Danilova L V, Pervouchine D D, Favorov A V, Mironov A A 2006 J. Bioinform. Comput. Biol. 4 589 DOI: 10.1142/S0219720006001904
|
| [38] |
Hofacker I L, Flamm C, Heine C, Wolfinger M T, Scheuermann G, Stadler P F 2010 RNA 16 1308 DOI: 10.1261/rna.2093310
|
| [39] |
Xayaphoummine A, Bucher T, Thalmann F, Isambert H 2003 Proc. Natl. Acad. Sci. 100 15310 DOI: 10.1073/pnas.2536430100
|
| [40] |
Xayaphoummine A, Bucher T, Isambert H 2005 Nucleic Acids Res. 33 605 DOI: 10.1093/nar/gki166
|
| [41] |
Gong S, Wang Y J, Zhang W B 2015 J. Chem. Phys. 142 015103 DOI: 10.1063/1.4905214
|
| [42] |
Zhao J, Hyman L, Moore C 1999 Microbiol. Mol. Biol. Rev. 63 405 DOI: 10.1128/MMBR.63.2.405-445.1999
|
| [43] |
Zhao P N, Zhang W B, Chen S J 2011 J. Chem. Phys. 135 245101 DOI: 10.1063/1.3671644
|
| [44] |
Gong S, Wang Y L, Wang Z, Wang Y J, Zhang W B 2018 J. Theor. Biol. 439 152 DOI: 10.1016/j.jtbi.2017.12.007
|
| [45] |
Chen J W, Zhang W B 2012 J. Chem. Phys. 137 225102 DOI: 10.1063/1.4769821
|
| [46] |
Chen J W, Gong S, Wang Y J, Zhang W B 2014 J. Chem. Phys. 140 025102 DOI: 10.1063/1.4861037
|
| [47] |
Wang Y L, Wang Z, Liu T G, Gong S, Zhang W B 2018 RNA 24 1229 DOI: 10.1261/rna.065961.118
|
| [48] |
Gong S, Wang Y J, Wang Z, Wang Y L, Zhang W B 2016 J. Phys. Chem. B 120 12305 DOI: 10.1021/acs.jpcb.6b09698
|
| [49] |
Colizzi F, Bussi G 2012 J. Am. Chem. Soc. 134 5173 DOI: 10.1021/ja210531q
|
| [50] |
Xu X, Yu T, Chen S J 2016 Proc. Natl. Acad. Sci. USA 113 116 DOI: 10.1073/pnas.1517511113
|
| [51] |
Wang Y J, Wang Z, Wang Y L, Zhang W B 2017 Chin. Phys. B 26 128705 DOI: 10.1088/1674-1056/26/12/128705
|
| [52] |
Wang Y J, Gong S, Wang Z, Zhang W B 2016 J. Chem. Phys. 144 115101 DOI: 10.1063/1.4944067
|
| [53] |
Wang Y J, Liu T G, Yu T, Tan Z J, Zhang W B 2020 RNA 26 470 DOI: 10.1261/rna.073882.119
|
| [54] |
Wang Y J, Wang Z, Wang Y L, Liu T G, Zhang W B 2018 J. Chem. Phys. 148 045101 DOI: 10.1063/1.5013282
|
| [55] |
Zhang W B, Chen S J 2006 Biophys. J. 90 765 DOI: 10.1529/biophysj.105.062935
|
| [56] |
Zhao P N, Zhang W B, Chen S J 2010 Biophys. J. 98 1617 DOI: 10.1016/j.bpj.2009.12.4319
|
| [57] |
Xia T, SantaLucia J, Burkard M E, Kierzek R, Schroeder S J, Jiao X, Cox C, Turner D H 1998 Biochemistry 37 14719 DOI: 10.1021/bi9809425
|
| [58] |
Urban S, Bartenschlager R, Kubitz R, Zoulim F 2014 Gastroenterology 147 48 DOI: 10.1053/j.gastro.2014.04.030
|
| [59] |
Diegelman-Parente A, Bevilacqua P C 2002 J. Mol. Biol. 324 1 DOI: 10.1016/S0022-2836(02)01027-6
|
| [60] |
Chadalavada D M, Senchak S E, Bevilacqua P C 2002 J. Mol. Biol. 317 559 DOI: 10.1006/jmbi.2002.5434
|
| [61] |
Macnaughton T B, Shi S T, Modahl L E, Lai M M C 2002 J. Virol. 76 3920 DOI: 10.1128/JVI.76.8.3920-3927.2002
|
| [62] |
Chadalavada D M, Knudsen S M, Nakano S, Bevilacqua P C 2000 J. Mol. Biol. 301 349 DOI: 10.1006/jmbi.2000.3953
|
| [63] |
Woodson S A, Emerick V L 1993 Mol. Cell. Biol. 13 1137 DOI: 10.1128/MCB.13.2.1137
|
| [64] |
Cao Y, Woodson S A 2000 RNA 6 1248 DOI: 10.1017/S1355838200000893
|
| [65] |
Chadalavada D M, Cerrone-Szakal A L, Bevilacqua P C 2007 RNA 13 2189 DOI: 10.1261/rna.778107
|
| [66] |
Delfosse V, Bouchard P, Bonneau E, Dagenais P, Centre-ville S 2010 Nucleic Acids Res. 38 2057 DOI: 10.1093/nar/gkp1080
|
| [67] |
Hennelly S P, Novikova I V, Sanbonmatsu K Y 2013 Nucleic Acids Res. 41 1922 DOI: 10.1093/nar/gks978
|
| [68] |
Perdrizet G A, Artsimovitch I, Furman R, Sosnick T R, Pan T 2012 Proc. Natl. Acad. Sci. USA 109 3323 DOI: 10.1073/pnas.1113086109
|
| [69] |
Mellin J R, Koutero M, Dar D, Nahori M-A, Sorek R, Cossart P 2014 Science 345 940 DOI: 10.1126/science.1255083
|
| [70] |
Feng J, Walter N G, Brooks C L 2011 J. Am. Chem. Soc. 133 4196 DOI: 10.1021/ja110411m
|
| [71] |
Kierzek E, Kierzek R 2020 J. Biol. Chem. 295 2568 DOI: 10.1074/jbc.H120.012787
|
| [72] |
Strobel B, Spöring M, Klein H, Blazevic D, Rust W, Sayols S, Hartig J S, Kreuz S 2020 Nat. Commun. 11 714 DOI: 10.1038/s41467-020-14491-x
|
| [73] |
Weinberg Z, Wang J X, Bogue J, Yang J, Corbino K, Moy R H, Breaker R R 2010 Genome Biol. 11 R31 DOI: 10.1186/gb-2010-11-3-r31
|
| [74] |
Breaker R R 2012 Cold Spring Harb. Perspect. Biol. 4 1 DOI: 10.1101/cshperspect.a003566
|
| [75] |
Li S, Hwang X Y, Stav S, Breaker R R 2016 RNA 22 530 DOI: 10.1261/rna.054890.115
|
| [76] |
Studer S M, Joseph S 2006 Mol. Cell 22 105 DOI: 10.1016/j.molcel.2006.02.014
|
| [77] |
Lin J C, Yoon J, Hyeon C, Thirumalai D 2015 Methods in Enzymology San Diego Elsevier Inc. 235 258 DOI: 10.1038/nrg2749
|
| [78] |
Reining A, Nozinovic S, Schlepckow K, Buhr F, Fürtig B, Schwalbe H 2013 Nature 499 355 DOI: 10.1038/nature12378
|
| [79] |
Wachter A, Tunc-Ozdemir M, Grove B C, Green P J, Shintani D K, Breaker R R 2007 Plant Cell 19 3437 DOI: 10.1105/tpc.107.053645
|
| [80] |
Cheah M T, Wachter A, Sudarsan N, Breaker R R 2007 Nature 447 497 DOI: 10.1038/nature05769
|
| [81] |
Lin J C, Thirumalai D 2013 J. Am. Chem. Soc. 135 16641 DOI: 10.1021/ja408595e
|
| [82] |
Rieder R, Lang K, Graber D, Micura R 2007 ChemBioChem 8 896 DOI: 10.1002/(ISSN)1439-7633
|
| [83] |
Lu C, Smith A M, Ding F, Chowdhury A, Henkin T M, Ke A 2011 J. Mol. Biol. 409 786 DOI: 10.1016/j.jmb.2011.04.039
|
| [84] |
Huang W, Kim J, Jha S, Aboul-ela F 2012 J. Mol. Biol. 418 331 DOI: 10.1016/j.jmb.2012.02.019
|
| [85] |
Sun T, Zhao C, Chen S J 2018 J. Phys. Chem. B 122 7484 DOI: 10.1021/acs.jpcb.8b04249
|
| [86] |
Ottink O M, Rampersad S M, Tessari M, Zaman G J R, Heus H A, Wijmenga S S 2007 RNA 13 2202 DOI: 10.1261/rna.635307
|
| [87] |
Soto A M, Misra V, Draper D E 2007 Biochemistry 46 2973 DOI: 10.1021/bi0616753
|
| [88] |
Tan Z J, Chen S J 2011 Biophys. J. 101 176 DOI: 10.1016/j.bpj.2011.05.050
|
| [89] |
Tan Z J, Chen S J 2010 Biophys. J. 99 1565 DOI: 10.1016/j.bpj.2010.06.029
|
| [1] | Chengwei Deng(邓成伟), Yunxin Tang(唐蕴芯), Jian Zhang(张建), Wenfei Li(李文飞), Jun Wang(王骏), and Wei Wang(王炜). RNAGCN: RNA tertiary structure assessment with a graph convolutional network[J]. 中国物理B, 2022, 31(11): 118702-118702. |
| [2] | Wei-Bu Wang(王韦卜), Xing-Yuan Li(李兴元), and Ji-Guo Su(苏计国). Force-constant-decayed anisotropic network model: An improved method for predicting RNA flexibility[J]. 中国物理B, 2022, 31(6): 68704-068704. |
| [3] | Yan-Hui Li(李彦慧), Zhen-Sheng Zhong(钟振声), and Jie Ma(马杰). Single-molecule mechanical folding and unfolding kinetics of armless mitochondrial tRNAArg from Romanomermis culicivorax[J]. 中国物理B, 2021, 30(10): 108203-108203. |
| [4] | Wen-Jing Wang(王文静) and Ji-Guo Su(苏计国). Enhancements of the Gaussian network model in describing nucleotide residue fluctuations for RNA[J]. 中国物理B, 2021, 30(5): 58701-058701. |
| [5] | . [J]. 中国物理B, 2021, 30(2): 28705-0. |
| [6] | Bin Huang(黄斌), Yuanyang Du(杜渊洋), Shuai Zhang(张帅), Wenfei Li(李文飞), Jun Wang(王骏), Jian Zhang(张建). [J]. 中国物理B, 2020, 29(10): 108704-. |
| [7] | Zhi-Chao Liu(刘志超), Qin Liu(刘琴), Chan-You Chen(陈禅友), Chen Zeng(曾辰), Peng Ran(冉鹏), Yun-Jie Zhao(赵蕴杰), Lei Pan(潘磊). [J]. 中国物理B, 2020, 29(10): 108709-. |
| [8] | Huiwen Wang(王慧雯), Yunjie Zhao(赵蕴杰). [J]. 中国物理B, 2020, 29(10): 108708-. |
| [9] | 何小玲, 王军, 王剑, 肖奕. Improving RNA secondary structure prediction using direct coupling analysis[J]. 中国物理B, 2020, 29(7): 78702-078702. |
| [10] | 杨毅, 辜琦, 张本龚, 时亚洲, 邵志刚. A novel knowledge-based potential for RNA 3D structure evaluation[J]. 中国物理B, 2018, 27(3): 38701-038701. |
| [11] | 熊贵, 席昆, 张曦, 谭志杰. To what extent of ion neutralization can multivalent ion distributions around RNA-like macroions be described by Poisson-Boltzmann theory?[J]. 中国物理B, 2018, 27(1): 18203-018203. |
| [12] | 王宇杰, 王珍, 王晏莉, 张文炳. Molecular dynamic simulation of the thermodynamic and kinetic properties of nucleotide base pair[J]. 中国物理B, 2017, 26(12): 128705-128705. |
| [13] | 常晓雪, 徐留芳, 史华林. Theoretical studies on sRNA-mediated regulation in bacteria[J]. 中国物理B, 2015, 24(12): 128703-128703. |
| [14] | 时亚洲, 吴园燕, 王凤华, 谭志杰. RNA structure prediction:Progress and perspective[J]. 中国物理B, 2014, 23(7): 78701-078701. |
|
||