|
Xiong Ling1, Liu Yanping1, 2, Liu Ruchuan1, Yuan Wei3, Wang Gao1, He Yi1, Shuai Jianwei2, Jiao Yang4, Zhang Xixiang5, Han Weijing6, Qu Junle3, ‡, Liu Liyu1, §
|
|
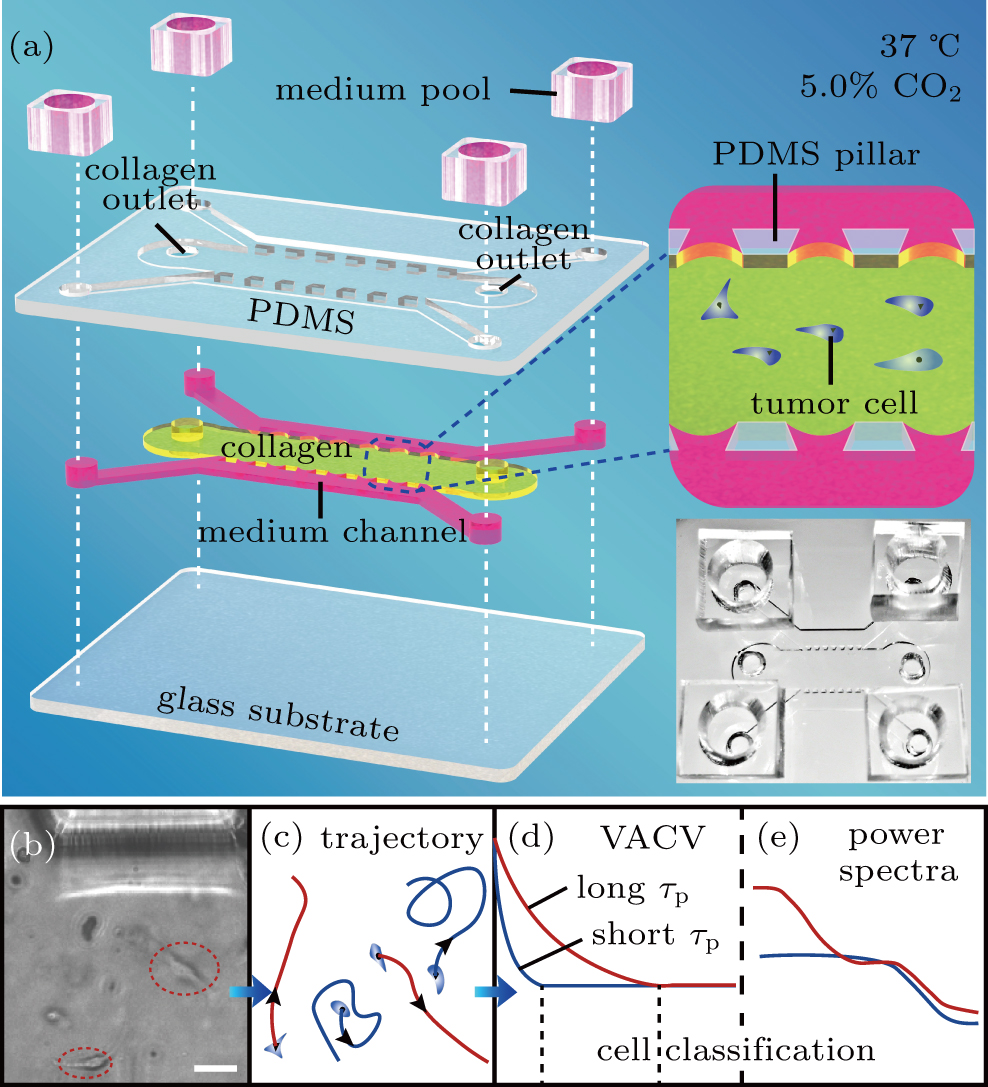
The microfluidic chip structure and cell classification steps. (a) Structure hierarchical diagram of the polydimethylsiloxane (PDMS) microfluidic chips. The PDMS scaffold (top layer) is bonded to a glass substrate (bottom layer), forming three channels (middle layer) separated by two arrays of micro-sized pillars (upper sides 200-
μ
m
long, bottom side 400-
μ
m
long, and high
200
μ
m) that center spacing 400
μ
m. Tumor cells are injected into the middle channel (7200-
μ
m
long and 3000-
μ
m
wide) with a collagen solution, which gelatinizes after incubation for 30 min at 37 °C and 5.0% CO2. The two shoulder channels are supplied with culture medium pools in experiments. This is also illustrated in the middle-right insert. Bottom-right insert displays a representative photo of a microfluidic chip. (b) and (c) Images of cell movement in collagen (scale bar,
50
μ
m
) and trajectory diagram, respectively. (d) and (e) Cell classification process diagram, based on VACV with different persistent times (
τ
p
) of cells and power spectra, respectively.
|