Improved data analysis method of single-molecule experiments based on probability optimization
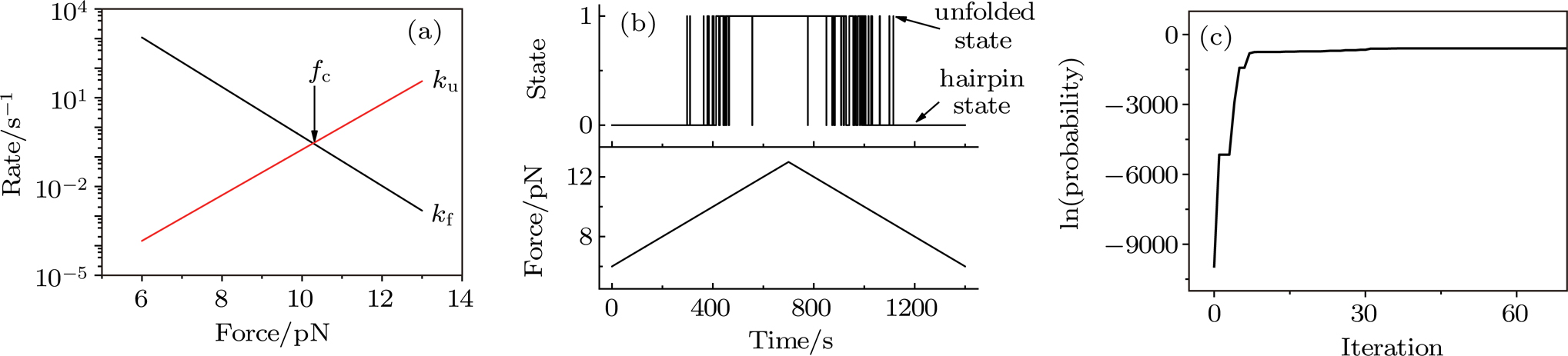
(color online) Simulation of transition at constant loading rate and optimization method. (a) Force-dependent folding and unfolding rates (Eqs. 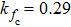
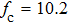
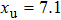
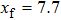
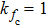
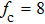
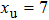
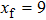
Improved data analysis method of single-molecule experiments based on probability optimization |
|
(color online) Simulation of transition at constant loading rate and optimization method. (a) Force-dependent folding and unfolding rates (Eqs. |
 |