† Corresponding author. E-mail:
Project supported by the Joint Funds of Xinjiang Natural Science Foundation, China (Grant No. 2015211C298).
Using a molecular theory, we investigate the temperature-dependent self-assembly of single-stranded DNA (ssDNA) tethered to a charged nanoparticle surface. Here the size, conformations, and charge properties of ssDNA are taken into account. The main results are as follows: i) when the temperature is lower than the critical switching temperature, the ssDNA will collapse due to the existence of electrostatic interaction between ssDNA and charged nanoparticle surface; ii) for the short ssDNA chains with the number of bases less than 10, the switching of ssDNA cannot happen, and the critical temperature does not exist; iii) when the temperature increases, the electrostatic attractive interaction between ssDNA and charged nanoparticle surface becomes weak dramatically, and ssDNA chains will stretch if the electrostatic attractive interaction is insufficient to overcome the elastic energy of ssDNA and the electrostatic repulsion energy. These findings accord well with the experimental observations. It is predicted that the switching of ssDNA will not happen if the grafting densities are too high.
Recently, much attention has been paid to developing the stimuli-triggered nanocarriers that can effectively regulate drug release in response to a given stimulus (e.g., a change in pH, or in temperature, or in magnetic field).[1–8] A DNA-based gatekeeper, as a potential candidate, provides new possibilities for the controlled drug delivery due to easy synthesis, high programmability, good biocompatibility, and efficient cellular uptake. In many applications, it is desirable to use solid surfaces as templates to guide DNA self-assembly or to maintain the stability of DNA-based nanostructures near surfaces.[9–12]
It is well established that DNA molecules are negatively charged in a solution under physiological conditions (pH ≈ 7). Many efforts have been made to exploit DNA-gated nanocarriers for the controlled release upon exposure to a specific pH, enzyme action, and near-infrared light.[9] A recent experiment on temperature-triggered DNA-gated nanocarrier performed by Yu et al.,[13] demonstrates that, when carboxyl-modified single-stranded DNA (ssDNA) is anchored to the positively charged nanoparticle surface, the state of ssDNA is directly controlled by changing body temperature. According to the experimental results of Ref. [13], the critical release temperature of nanocarriers is with the DNA value of N > 10 (N is the number of bases). The sequences of 10, 20, 30, and 40 bases long ssDNA are 5’-COOH-(CH2)6-ACTCCTGGTA-3’, 5’-COOH-(CH2)6-ACTCCTGGTATGTAGCGCTA-3’, 5’-COOH-(CH2)6-ACTCCTGGTATGTAGCGCTAACTGTAATGG-3’, and 5’-COOH-(CH2)6–ACTCCTGGTATGTAGCGCTAACTGTAA TGGTCACGTCACT-3’, respectively.[13] Nevertheless, the cargo molecules are released continuously at all the set temperatures if the length of the DNA valve is decreased to 10 bases. The experimental results imply that, for a 20–40 base long ssDNA chain, grafted ssDNA will stretch at high temperatures, and will collapse at low temperatures. For short ssDNA chains (N = 10), the temperature-dependent switching of ssDNA cannot happen. Understanding the behavior of DNA tethered to charged surface is needed to guide the above applications. Yamamoto et al.[14] predicted the electric-field-dependent collapse of a polyelectrolyte brush grafted on an electrode using a mean field theory. Using molecular dynamics simulations, Robert et al.[15] studied short ssDNA chains on model hydrophilic and hydrophobic self-assembled monolayers (SAMs), and elucidated the molecular interactions between the ssDNA, SAM, and water molecules. These ssDNA chains exhibit a favorable binding energy on both hydrophilic and hydrophobic SAMs. However, up to now, few theories have been devoted to a molecular-level understanding of temperature-dependent structure and switching of ssDNA tethered to a charged nanoparticle surface. Thus, it is necessary for us to include more molecular details to describe the structure and switching of ssDNA properly.
In this paper, motivated by the intriguing results of Ref. [13], we will present a molecular theory[16–20] to explore how charged surfaces influence the structure and switching of ssDNA chains, and to explain the mechanism for temperature-dependent switching of ssDNA. The remaining part of this paper is organized as follows: in Section 2, we describe a molecular model; in Section 3, we present the results of investigating the temperature-dependent structure and switching of ssDNA tethered to a charged nanoparticle surface; we give conclusions and discussion in Section 4.
Amino-modified mesoporous silica nanoparticles (MSNs-NH2) were selected.[13] MSNs-NH2 was positively charged. Carboxyl-modified ssDNA was anchored to the amino group on the surface of MSNs-NH2.[13] The system we consider is composed of ssDNA-grafted charged nanoparticles immersed in water. The nanoparticle with a radius R = 37 nm has its center at the origin of the r axis, and its surface is grafted by ssDNA. Carboxyl will not significantly affect the ssDNA grafting density. The grafting density is dependent on grafting temperatures, grafting reaction time, and molecular weight.[21,22] The number of grafted chains per unit area, or grafting density, is σ = Np/4πR2. Our model ssDNA is a long chain with N segments (i.e., N bases), and each segment has a volume of vp = 0.4 nm3.[23] Water, proton, and hydroxyl ions are assumed to have the same volume of vw = 0.03 nm3. The existence of nanoparticles induces an inhomogeneous distribution of all the molecular species, and the only inhomogeneous direction is assumed to be along the r axis. Considering the size, shape, and conformation of every molecular type explicitly, the Helmholtz free energy of the system is given by

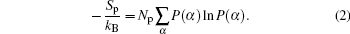
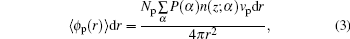
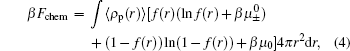
The third term on the right side of Eq. (
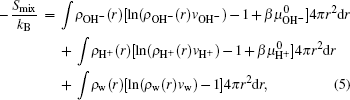


The fourth term on the right side of Eq. (
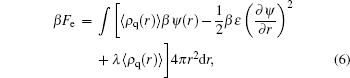
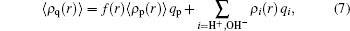
The fifth term on the right side of Eq. (

The last term of Eq. (


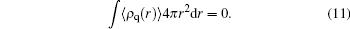
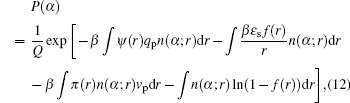
For the electrostatic potential, we obtain a generalized Poisson–Boltzmann equation



Equation (
The expression for the fraction of the charged polymer segments at r is

In a bulk solution, the acid–base equilibrium is determined by the equilibrium constant, 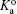

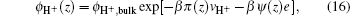
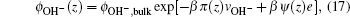
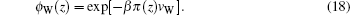
The bulk values of ions can be obtained from the ions concentration: ϕbulk,∓ = c∓Nav∓. The unknowns in the above equations are the position-dependent electrostatic and pressure fields. These quantities are determined by substituting Eqs. (
In this section, we present some representative results for the structure and switching of ssDNA tethered to the charged nanoparticle surface.
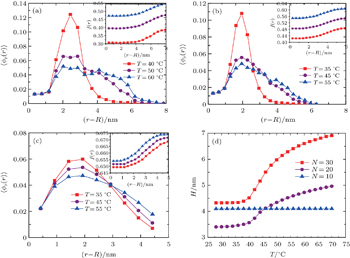
Figure
 | Fig. 2. Height of grafted ssDNA chains as a function of temperature. The number of segments of grafted ssDNA chains is N = 40. The grafting density is σ = 0.5 nm−2. |
In order to understand the origin of this behavior, we analyze the switching of ssDNA tethered to the charged nanoparticle surface in terms of varying chain length and behavior of charged ssDNA chains, and produce the diagrams of local average volume fractions as a function of distance from the nanoparticle surface, as shown in Figs.
Figure
It is worthwhile to note that the screened Coulombic interactions give rise to a negative pressure, and counterions result in an entropic pressure, which can induce the collapse of the polyelectrolyte.[14] The counterion-mediated attractive interactions along a chain can also lead to a collapse of a single chain, endowing the chain with a very compact structure and an inhomogeneous distribution. To explore the screened Coulombic interactions and attractive interactions along chains, we investigate distributions of H+ and OH−, and produce the diagrams of local fractions of local concentration of H+ and OH− as a function of distance from the charged nanoparticle surface at different temperatures, as shown in Figs.
 | Fig. 5. Local concentration of (a) H+ and (b) OH− as a function of the distance from the nanoparticle surface. |
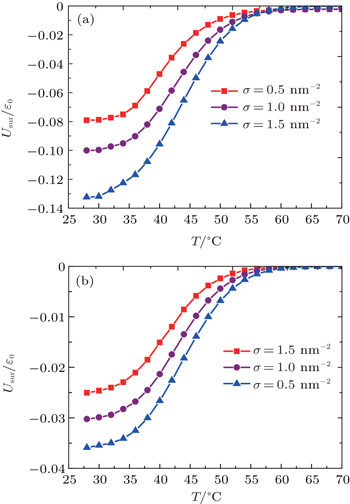
So far, we have discussed the temperature effect on the self-assembly of ssDNA chains. To further verify the structure and switching of ssDNA tethered to the charged nanoparticle surface, it is instructive to reflect on Eq. (
As illustrated in Fig.
For practical applications of DNA-gated nanocarriers,[13] grafting density must be controllable. Choosing different temperatures, we intend to see the impact of H on grafting density. Figure
 | Fig. 7. The height of the grafted ssDNA layer as a function of the grafting density for (a) N = 40 and (b) N = 10. |
In order to quantify the charging behavior of the ssDNA chains in response to changes in grafting density, we define the average fraction[27,28] of the charged polyelectrolyte monomers by

Figure
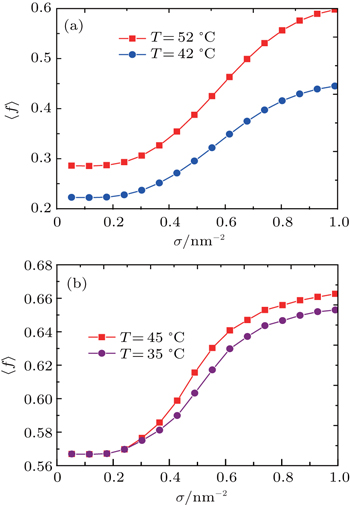 | Fig. 8. The average fraction of charged polyelectrolyte monomers as a function of grafting density for (a) N = 40 and (b) N = 10. |
Figure
To gain greater entropy and to reduce the energy cost from ssDNA intermolecular repulsions, it will be favorable for many monomers to remain charged. However, most ssDNA molecules in short ssDNA chains (N = 10) are charged. Thus, ⟨f⟩ represents slightly nonmonotonic change in response to increasing grafting density, as shown in Fig.
We have employed a molecular theory to study the temperature-dependent structure and switching of ssDNA tethered to a charged nanoparticle surface. For the longer (N > 10) ssDNA chains, they adopt a compact conformation on the bottom of a layer at low temperatures, while the inverse transformation will take place at high temperatures. The electrostatic attractive interaction between the ssDNA and charged nanoparticle surface weakens as temperature increases. When the electrostatic attractive interaction cannot overcome the elastic energy of ssDNA and electrostatic repulsion energy, ssDNA chains will stretch. For the short (N = 10) ssDNA chains, the switching of ssDNA cannot happen, and the critical temperature does not exist. This is because, in all ranges of temperature, the electrostatic attraction interaction between the negatively charged polymer segments and the positively charged surface is insufficient to overcome the elastic energy of ssDNA and electrostatic repulsion energy along ssDNA chains. This accords well with the experimental observations[13] of ssDNA at different temperatures, and reveals the unique temperature-dependent structure and switching behavior of the ssDNA tethered to a charged nanoparticle surface.[13] According to our theoretical model, we can predict that if the grafting densities are too high, the switching of ssDNA cannot happen.
Our theoretical approach has general characteristics, which will be applied to other molecule systems, such as single-stranded PNA and single-stranded RNA.[31–36] These molecule systems have a great potential for applications in biomaterials. It is worth mentioning that experiments in previous studies were carried out under nonequilibrium conditions. However, we believe that main temperature-dependent switching of ssDNA tethered to a charged nanoparticle surface is captured by our equilibrium approach. The slow non-equilibrium kinetics, as observed in the experiments of Ref. [13], can lead to lateral phase separation,[37] making the ssDNA tethered to charged nanoparticle surface density nonhomogenous. It is necessary to build a theoretical model by taking into account a three-dimensional nonequilibrium system. Advances in theory,[38–43] combined with experimental observations, will enable us to modify the designs of biopolymer surfaces for a large variety of applications.
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 | |
| 10 | |
| 11 | |
| 12 | |
| 13 | |
| 14 | |
| 15 | |
| 16 | |
| 17 | |
| 18 | |
| 19 | |
| 20 | |
| 21 | |
| 22 | |
| 23 | |
| 24 | |
| 25 | |
| 26 | |
| 27 | |
| 28 | |
| 29 | |
| 30 | |
| 31 | |
| 32 | |
| 33 | |
| 34 | |
| 35 | |
| 36 | |
| 37 | |
| 38 | |
| 39 | |
| 40 | |
| 41 | |
| 42 | |
| 43 |