† Corresponding author. E-mail:
Project supported by the National Natural Science Foundation of China (Grant No. 11205186).
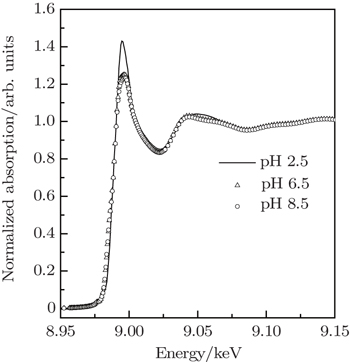
The local configurations around metal ions in metalloproteins are of great significance for understanding their biological functions. Cu2+/histidine (His) is a typical complex existing in many metalloproteins and plays an important role in lots of physiological functions. The three-dimensional (3D) structural configurations of Cu2+/His complexes at different pH values (2.5, 6.5, and 8.5) are quantitatively determined by x-ray absorption near-edge structure (XANES). Generally Cu2+/His complex keeps an octahedral configuration consisting of oxygen atoms from water molecules and oxygen or nitrogen atoms from histidine molecules coordinated around Cu2+. It is proved in this work that the oxygen atoms from water molecules, when increasing the pH value from acid to basic value, are gradually substituted by the Ocarboxyl, Nam, and Nim from hisitidine molecules. Furthermore, the symmetries of Cu2+/His complexes at pH 6.5 and pH 8.5 are found to be lower than at pH 2.5.
Complexes of transition metals and amino acid residues play important roles in many biological processes, such as electron transfer, redox and catalysis.[1] In these processes, the enzyme active sites are highly specific and often involve histidine (His) residues interacting with divalent transition metal cations, i.e., Mn2+, Fe2+, Co2+, Ni2+, Cu2+, and Zn2+.[2] Furthermore, these metals are the essential trace elements for living bodies. The deficiencies of them will cause diseases and a supplementation is needed. Typically, the deficiency of Cu will cause the Menkes disease, a rare genetic disorder. Subcutaneous administration of the Cu2+/His complex is the only promising way to treat this disease.[3–6] To study the function of Cu2+/His complex in Cu transport, it has already been characterized by many physical techniques, such as infrared, Raman, extended x-ray absorption fine structure (EXAFS) since it was discovered in human blood in 1966.[7,8]
The molecule formula of His is C6H9N3O2, including one imidazole ring. There are several potential coordination sites for metal ions to coordinate the His molecule, e.g. the N atom from amino (Nam), the O atom from carboxyl (Ocarboxyl) and the two N atoms from imidazole (they are both named Nim, the one near Cβ is Nπ, the other one is Nτ). So the structure of Cu2+/His complex is complicated. The results obtained by other work suggest that Cu2+/His complex keeps an octahedral configuration with O/N atoms from water molecules and His coordinated with Cu ion.[8] The details regarding the three-dimensional (3D) local structure of Cu2+/His in solutions with different pH values was not yet investigated.
X-ray absorption fine structure (XAFS) is a powerful technique to detect the local structure information about metal ions at high spatial resolution, which has been successfully used in lots of research fields.[9–15] It is composed of two parts, i.e., EXAFS and x-ray absorption near-edge structure (XANES). XANES is more sensitive to the local structure configuration and coordination atoms around metal atoms than EXAFS. In order to make a further study, the structures of Cu2+/His complexes at three typical pH values (2.5, 6.5, and 8.5) are studied by both qualitative and quantitative XANES analysis. In the quantitative analysis, MINUIT XANes (MXAN) that employs the full multiple scattering (MS) approach to a description of the XANES region was used.[16–19] MXAN has been proved to be powerful in a number of interesting situations.[20–24] It can be employed to reveal more detailed high resolution 3D structures of the studied systems with a lower command for S/N than EXAFS.
Solutions of Cu2+/His with a molar ratio of 1:2 at pH values of 2.5, 6.5, 8.5 were prepared by dissolving 0.01-Mol L-histidine and 0.005-Mol CuCl2·2H2O into the de-ionized water with a volume of 40 ml. Then NaOH or HCl solution was added to adjust the pH value.
XAFS measurements were performed at the XAFS station of 1W1B beamline of the Beijing Synchrotron Radiation Facility (BSRF). The typical energy of the storage ring was 2.5 GeV with a current decreasing from 250 mA to 160 mA during experiment runs. An Si (111) double crystal monochromator was used to minimize the high harmonics content through the detuning of the two crystals by 20%. The absolute energy position was calibrated using a Cu metal foil. The XAFS spectra of Cu K-edge of Cu2+/His at different pH values were recorded in the fluorescence mode. For the experiments, the solution sample was kept in a sample cell of polyethylene. An ion chamber which 50% argon and 50% nitrogen flowed into was used to monitor the incident beam intensity (I0) while a Lytle detector which pure Ar flowed into was used to record the fluorescence signal (If). Data of all the experimental XAFS spectra were analyzed by using WinXAS3.1[25] and the MXAN package.[16–19] The initial models of Cu2+/His complexes at different pH values for XANES analysis were established according to the results suggested by other works.[8]
The XANES spectra at Cu K-edge of Cu2+/His complexes at pH 2.5, pH 6.5, and pH 8.5 are compared in Fig.
Quantitative XANES analyses of different samples obtained by MXAN and the corresponding structure sketches are shown in Figs.
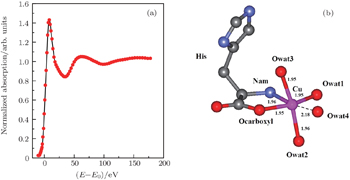 | Fig. 2. (a) XANES fitting results of Cu2+/His complex at pH 2.5, calculated by MXAN, and (b) the corresponding structure sketch. |
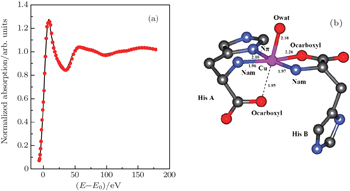 | Fig. 3. (a) XANES fitting results of Cu2+/His complex at pH 6.5, calculated by MXAN, and (b) the corresponding structure sketch. |
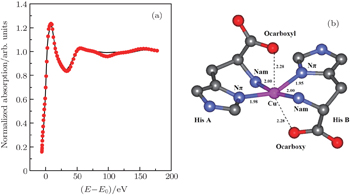 | Fig. 4. (a) XANES fitting results of Cu2+/His complex at pH 8.5, calculated by MXAN and (b) the corresponding structure sketch. |
| Table 1. Values of bond length (Å) between Cu ion and the first shell atoms (O/N) from water molecules and His of Cu2+/His complex at pH 2.5, calculated by MXAN. . |
| Table 2. Angles (°) between the different coordination sites (ligands) of Cu2+/His complex at pH 2.5, calculated by MXAN. In the table the definitions of the ligands are as follows: Ligand0: Cu2+; Ligand1: histidine (His); Ligand2: water molecule 1 in the horizontal plane (wat1); Ligand3: water molecule 2 on axis (wat2); Ligand4: water molecule 3 on axis (wat3); Ligand5: water molecule 4 in the horizontal plane (wat4). . |
| Table 3. Values of bond length (Å) between Cu ion and the first shell atoms (O/N) from water and His of Cu2+/His complex at pH 6.5, calculated by MXAN. . |
| Table 4. Angles (°) between the different coordination sites of Cu2+/His complex at pH 6.5, calculated by MXAN. In the table, the definitions of the ligands are as follows: Ligand0: Cu2+; Ligand1: histidine A (His A); Ligand2: histidine B (His B); Ligand3: water molecule on axis (wat). . |
| Table 5. Values of bond length (Å) between Cu ion and the first shell atoms (O/N) from His of Cu2+/His complex at pH 8.5, calculated by MXAN. . |
| Table 6. Angles (°) between the different coordination sites of Cu2+/His complex at pH 8.5, calculated by MXAN. In the table, the definitions of the ligands are as follows: Ligand0: Cu2+; Ligand1: histidine A (His A); Ligand2: histidine B (His B). . |
The stereo structures of Cu2+/His complexes at different pH values are quantitatively determined by XANES at the Cu K-edge. From the XANES spectra, it is confirmed that Cu2+/His complex maintains an octahedral configuration with O/N atoms from water molecules and His molecules. Nevertheless, the local geometric structure varies with the pH value of the solution environment. Firstly, at pH 2.5, i.e., under the acid condition, only two atoms (Ocarboxyl and Nam) from one His molecule are coordinated with the Cu ion while the O atoms from water molecules occupy the other four coordination sites. The Cu-His bond length is mainly about 1.95 (1.96) Å and the molar ratio between Cu2+ and His is 1:1. As the pH value increases to 6.5, the number of atoms from the His molecule which are coordinated with the Cu ion, increases to 5, including Ocarboxyl, Nam, and Nπ from two His molecules; while the oxygen coordinates from water decrease to one along the axis. The Cu-His bond length is not equally distributed. Of the bond lengths, the shortest length is about 1.95 Å and the longest length is 2.20 Å. The molar ratio between Cu2+ and His is 1:2. Finally, as the pH value increases up to 8.5, i.e., basic condition, the coordinates of Cu ion are purely from two His molecules, each of which has three coordinating atoms, i.e., Ocarboxyl, Nam, and Nπ. The bond length increases slightly to 1.98 Å and 2.28 Å. The molar ratio of Cu2+ and His remains 1:2. It is concluded that Ocarboxyl, Nam, and Nπ from His molecules substitute for O atoms from water molecules in Cu2+/His complex gradually when the pH value of the solution increases from acid (pH 2.5) to basic (pH 8.5), and they have a priority to be coordinated with the Cu ion compared with the Nτ atom. Furthermore, it also shows that the bond lengths of the first shell atoms in Cu2+/His complexes at pH 6.5 and pH 8.5 are not as equally displaced as that at pH 2.5, pointing out a lower symmetry of Cu2+/His in basic solution than in acid solutions, which is consistent with the qualitative analysis of XANES.
In this work, the local structural configurations of Cu2+/His complexes at different pH values are performed by using the XANES spectroscopy. The 3D stereo structural information is extracted by fitting the near-edge spectra. The geometric structure becomes disordered in basic solution, resulting in the variations of coordinates and bond distances. This work is not only complementary to a general understanding of the structure in Cu2+/His complex, but also facilitates our understanding of the physiological mechanisms of Cu2+/His in biological systems.
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 | |
| 10 | |
| 11 | |
| 12 | |
| 13 | |
| 14 | |
| 15 | |
| 16 | |
| 17 | |
| 18 | |
| 19 | |
| 20 | |
| 21 | |
| 22 | |
| 23 | |
| 24 | |
| 25 |