† Corresponding author. E-mail:
Project supported by the National Natural Science Foundation of China (Grant No. 21403182) and the Research Grants Council of Hong Kong, China (Grant No. CityU 21300014).
Membrane curvature is no longer thought of as a passive property of the membrane; rather, it is considered as an active, regulated state that serves various purposes in the cell such as between cells and organelle definition. While transport is usually mediated by tiny membrane bubbles known as vesicles or membrane tubules, such communication requires complex interplay between the lipid bilayers and cytosolic proteins such as members of the Bin/Amphiphysin/Rvs (BAR) superfamily of proteins. With rapid developments in novel experimental techniques, membrane remodeling has become a rapidly emerging new field in recent years. Molecular dynamics (MD) simulations are important tools for obtaining atomistic information regarding the structural and dynamic aspects of biological systems and for understanding the physics-related aspects. The availability of more sophisticated experimental data poses challenges to the theoretical community for developing novel theoretical and computational techniques that can be used to better interpret the experimental results to obtain further functional insights. In this review, we summarize the general mechanisms underlying membrane remodeling controlled or mediated by proteins. While studies combining experiments and molecular dynamics simulations recall existing mechanistic models, concurrently, they extend the role of different BAR domain proteins during membrane remodeling processes. We review these recent findings, focusing on how multiscale molecular dynamics simulations aid in understanding the physical basis of BAR domain proteins, as a representative of membrane-remodeling proteins.
The formation of intracellular membrane compartments allows the cell to enable various coordinations for the numerous biochemical reactions required in eukaryotic life. While this event takes place when phospholipids become abundant enough to form the barriers surrounding the cell or to form its intracellular organelles, communication between the “inside” and “outside” of the cell requires interplay between the lipid bilayers and cytosolic proteins. This communication, is usually achieved via a multitude of tiny membrane bubbles known as vesicles, which act as carriers. Flux of these vesicles, that is, trafficking, not only contributes to influx of nutrients and secretion of wastes but is also strongly associated with well-known diseases such as cancer, Alzheimer’s disease (AD), and other neurodegenerative diseases. [ 1 – 3 ] Studying the detailed process of how proteins initiate the formation of membrane curvature, sculpt a vesicle of defined size, and pinch off the new bud is therefore not simply a matter of scientific curiosity but is also directed toward new breakthroughs in modern physiology.
Given that the first visual proof for curvature-generating proteins was published in 1964 [ 4 ] and was followed by reports of distinct membrane-bending proteins such as the Bin/Amphiphysin/Rvs (BAR) domain proteins or the epsin N-terminal homology (ENTH) domain proteins, [ 5 , 6 ] study of the mechanism underlying membrane curvature induction by proteins has progressed rather slowly. This is attributable to the difficulty experienced in direct visualisation and investigation of the dynamics of curvature generation. While a breakthrough in technology always gives rise to breakthroughs in science; the emergence of high-resolution structures of membrane-remodeling proteins [ 7 – 9 ] not only helps in closing the knowledge gap but also transforms the subject into a rapidly developing field because of interesting computational studies that provide experimentally unobtainable atomistic information on cellular mechanisms. As a result, crystal structures and molecular dynamics (MD) simulations (and the combination of the two) have so far strongly influenced our understanding of membrane bending by peripheral proteins.
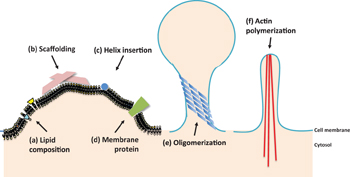
This review will start by introducing several known mechanisms underlying membrane curvature induction (Fig.
There are several known mechanisms of membrane curvature induction (Fig.
The effect of lipid composition on membrane curvature originates from the intrinsic shape of different lipid headgroups or acyl chains. Side-by-side packing of lipids will impose curvature on the monolayer according to the shape of the local lipids. Generally, lipids with a small headgroup and large acyl chains, i.e., unsaturated tails, prefer negative curvatures, while lipids with a large headgroup and saturated tails prefer positive curvatures. Owing to the coupling of the lipid bilayer, it was expected that a greater portion of lipids with positive preferred curvature would be found in outer monolayers and those with negative preferred curvature would be found in inner monolayers during vesicle budding. [ 20 ] This was concurrently proven by the observation that highly curved regions selectively include or exclude specific lipids on the basis of their shape or stiffness and that a significant proportion of lipids with unsaturated tails is found in highly curved membrane tubes. [ 20 – 22 ] For examples, phosphatidylinositol phosphates (PIs) and lysophosphatidylcholine (LPC) with a large headgroup to acyl tail size ratio show inverted conical shapes and therefore favor intrinsic positive curvatures of the membrane. In contrast, phosphatidylethanolamine (PE), phosphatic acid, diacylglycerol (DAG) and cardiolipin have relatively small headgroups and thus a conical shape, leading to negative membrane curvatures of the monolayer. While the lipid does not have either too large or too small head-to-tail ratio, it is called a cylindrical lipid, for examples, phosphatidylcholine (PC) and phosphatidylserine (PS); these lipids are simply packed to form a flat monolayer. [ 22 – 24 ]
In addition to headgroup size, saturation of acyl chains also contributes to the headgroup to acyl tail size ratio. Kinks induced by double bonds obviously occupy more space in unsaturated tails than in saturated tails. For example, 1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS), which was created artificially, favors spontaneous curvature because of its bulky headgroup and monounsaturated acyl chains. [ 25 ] Polyunsaturated phospholipids (PLs) have been recently reported to increase the ability of dynamin and endophilin to deform and vesiculate synthetic membranes. [ 26 ]
Imposing the shape of the membrane-binding interface onto the membrane is another effective way of membrane remodeling and is often called as scaffolding (Fig.
BAR domains are crescent-shaped dimers that bind to membranes and will be the main focus of this review. As a major representative of peripheral membrane-scaffolding proteins, these domains were first noticed as the most conserved feature in amphiphysins from yeast to humans and also in endophilins and nadrins. [ 8 ] BAR domains can remodel the membrane according to their intrinsic banana shapes, a process that is mediated by electrostatic interactions between positively charged residues (lysine or aginine) on the concave binding surface and negatively charged lipids (PS and/or PI phosphates). [ 27 , 28 ] BAR domains were previously found to sense membrane curvatures as they bind more readily to liposomes whose curvature is closer to the intrinsic curvature of the BAR. [ 8 , 29 ] At a higher concentration, this sensor was thought to become an inducer. [ 30 ] The energetics of amphiphysin on the membrane also show a close relationship between the binding and bending energies during membrane remodeling. [ 31 ]
An additional feature of the BAR domains is their tight correlation with an N-terminal amphipathic helix (an N-BAR domain). As will be discussed in detail in Section 3, this amphipathic helix also causes membrane bending; many studies have been performed on cooperation between the BAR domain and the amphipathic helix. [ 8 , 32 , 33 ] Whether the scaffolding mechanism is responsible for the membrane curvature or just serves as a prerequisite for helix insertion is still under debate. In any case, such combination has frequently been noted in membrane remodeling proteins such as amphiphysin, endophilin, BRAP, and nadrin and all these have been reported to induce tubulation in vitro . [ 5 , 34 – 36 ]
Members of the dynamin family (which are also dimers) were also reported to scaffold inositol lipids, leading to tubulation of the membrane with formation of helical oligomers. [ 29 , 37 ] The dynamin protein is seldom discussed without mentioning BAR domains as they cooperate with each other to a great extent in many membrane remodeling events such as clathrin-mediated endocytosis. [ 5 , 38 ] In addition to simple attachment to the membrane, this family of proteins uses GTP hydrolysis to effect membrane fission (scission) which has been discussed in a few early studies. [ 29 , 37 , 39 ] It is important to acknowledge the role of different proteins that are involved in the scaffolding of membranes; the details of the fission process, however, are beyond the scope of this review.
Another effective way of inducing local membrane curvature is to insert a hydrophobic protein motif between the lipid headgroups. The motif acts like a wedge; when it is inserted near the lipid glycerol backbones, the adjacent hydrocarbon chains tilt and splay to fill the resultant vacant spots underneath the wedge [ 11 ] (Fig.
Sometimes the hydrophobic motif appears on amphipathic helices which are stretches of α -helix with one side being polar (charged) and the other hydrophobic. [ 11 , 30 ] Amphipathic helices are present in many proteins that participate in different stages of the membrane-remodeling process, such as endophilin, amphiphysin, epsin, nadrin, Arf proteins and the secretion-associated RAS-related 1 (SAR1) GTPase. [ 42 ] In the case of many of these proteins, the amphipathic helices are unfolded before they insert into membranes. As long as the helices insert into the membranes and fold, they sit flat on the surface with the hydrophobic residues dipping into the hydrophobic phase of the membrane. Another key feature of the amphipathic helices is the nature of their polar face, which grants them various abilities such as sensing membrane curvatures.
Other proteins such as the C2 domains of synaptotagmin-1 and Doc2b also insert hydrophobic loops that are much more bulky than a four-turn helix in order to induce greater curvatures. [ 41 , 43 – 45 ] The membrane-binding interface of PACSINs, which is a subfamily of the amino-terminal Bin–Amphiphysin–Rvs (F-BAR) domain, and EHDs, which comprise a class of eukaryotic ATPases involved in clathrin-independent endocytosis, also uses hydrophobic loops for insertion. [ 46 , 47 ] Similar to the amphipathic helices, the nature of these hydrophobic loops varies across the proteins. The membrane-bending activity of C2 domains may be regulated by the Ca 2+ concentration. [ 43 , 44 ]
Transmembrane proteins (commonly known as ion channels, transporters and receptors) that have an intrinsic conical or inverted conical shape can bend associated membranes due to asymmetry of extramembrane domains of the proteins. [ 14 – 18 ] Protein crowding might also occur for receptors where the lipid bilayer bends from the side with a larger domain. [ 48 ] Sometimes integral proteins even oligomerize to cause local scaffolding of membranes. [ 49 , 50 ] Clustering of transmembrane receptors, such as transferrin or low-density lipoprotein (LDL) receptors, was reported to aid in pit stabilization during clathrin-mediated endocytosis. [ 51 ] Such a crowding mechanism has been suggested to explain the binding of some proteins to the membrane surface at a relatively high concentration but the contribution of crowding to curvatures is still unclear. [ 52 , 53 ]
Completion of the process of vesicle budding under physiological conditions usually requires a combination of the aforementioned mechanisms. Such a cellular process involves proteins with scaffolding abilities, helix-insertion abilities, or both. Moreover, local curvatures induced by single domains, either by scaffolding by a concave surface or by insertion of a single helix, dissipate rapidly in space; proteins in cells, oligomerize to induce much more pronounced membrane curvatures (Fig.
BAR domain proteins are frequently reported to oligomerize in order to induce macroscopic membrane curvatures such as tubulation and vesiculation. [ 54 , 55 ] A series of both experimental and computational studies have been conducted for in-depth analysis of BAR oligomers and their contribution to membrane curvatures. [ 56 – 58 ]
Members of the dynamin family are reported to polymerize into spirals in order to remodel membranes. The fact that they require pre-existing curvatures to assemble efficiently and are GTP-dependent again identifies their main role of inducing scission events instead of early membrane sculpting. [ 59 – 62 ]
Some coat proteins are formed from oligomerization of single domains such as clathrin, COPI, and COPII, mainly for stabilizing existing membrane curvatures during vesicle budding. [ 63 , 64 ] Instead of binding directly, they rely on adaptor proteins in order to link to membranes; therefore, this process is sometimes classified as indirect scaffolding. The ability to bend the membranes depends on the rigidity of the assembled polyhedral coat and the transmission of the shape to the membrane. [ 48 ]
Budding of a clathrin-coated vesicle is a good example of the combination of membrane remodeling mechanisms. Inverted conical lipids (PS and PIP2) asymmetrically distribute between the two membrane monolayers and the intrinsic local curvature is stabilized or even reinforced by proteins with scaffolding or insertion modules; all this is mediated by an oligomeric protein scaffold enclosed by the clathrin coat. [ 13 ] As oligomerization represents the collective modes of one or more types of proteins, it is extremely complicated and introduces big challenges on both experimental and theoretical fronts.
Shaping of cellular organelles, which also requires global membrane curvatures employs protein oligomerization of caveolin, flotillins, or the reticulons. Similar to clathrin coats, these proteins sometimes utilize bridging proteins to either scaffold or insert motifs into membranes in order to facilitate the formation and stabilization of curvatures at caveolae and the endoplasmic reticulum (ER). [ 65 – 67 ]
Macroscopic membrane curvatures are present in large intracellular organelles, such as the Golgi or the ER. Scaffolding by actin, intermediate filament (Fig.
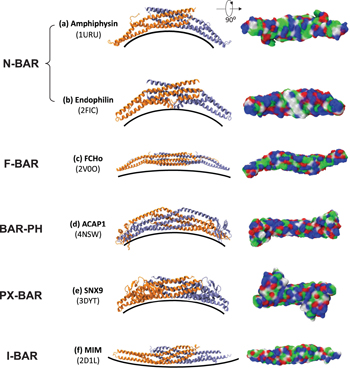
Among all the membrane-bending proteins, the superfamily of BAR proteins has become the main focus of attention over recent years. Numerous reports confirmed that these proteins participate in shaping membranes from yeast to mammals. [ 71 ] BAR family proteins commonly utilize the helical insertion or scaffolding mechanism to deform the membrane together with oligomerization. [ 28 , 72 ] Recently BAR proteins have even been reported to switch from wedging (helical insertion) to scaffolding depending on the protein concentration. [ 73 ] BAR domains are remarkably conserved at the structural level, featuring a three helix coiled coil core that forms curved dimers and results in the characteristic ‘banana shape’ (Fig.
BAR proteins are categorized based on the general type of their BAR domain: classical BAR domain, extended FCH domain (F-BAR), BAR domains with additional membrane-binding domains (BAR-PH and PX-BAR), or IMD/inverse BAR domain (I-BAR) (Fig.
Classical BAR domains, as found in arfaptin, were the first BAR domains to be discovered. [ 76 ] This type of BAR domains also includes BAR domains with an amphipathic N-terminal helix which are often named as N-BAR domains (e.g., amphiphysins and endophilins) (Figs.
F-BAR domain proteins, such as Cdc42-interacting protein 4 (CIP4), forming binding protein 17 (FBP17), and Fes-CIP4 homology domain (FCHO) (Fig.
Some BAR domains are found to have additional membrane binding domains such as the pleckstrin homology domain (BAR-PH, e.g., in APPL1 and ACAP1) (Fig.
I-BAR domains are currently the only subfamily of BAR domains having a negative curvature (Fig.
MD simulation is an important tool that provides knowledge at the atomic level of the structural and dynamic aspects of biological systems for understanding the physical basis of these complex molecular assemblies. The first MD simulation of a protein was published more than 30 years ago, [ 84 ] and credited to the availability of relatively accurate x-ray structures of macromolecule in 1975. With the rapid developments in novel experimental techniques, limits to the resolution, size, and complexity of the biological systems that can be investigated through MD simulations have been constantly extended. [ 85 ] The availability of more sophisticated experimental data poses challenges to the theoretical community for developing novel theoretical and computational techniques that can be used to better interpret the experimental results for further functional insights. The simulation methodology utilized in a number of widely used programs originally developed in the 1980s (CHARMM [ 86 ] and AMBER [ 87 ] ) is constantly being improved further. Optimization of the internal parameters for the all-atom empirical energy function in the programs used experimental gas-phase geometries, vibrational spectra, and torsional energy surfaces supplemented with ab initio results. [ 88 ] Developments in theoretical models continue apace with applications of MD simulations and, at the same time, benefit from improvements in the speed of computers. Until now, highly efficient parallel MD simulation codes (such as NAMD [ 89 ] and Gromacs [ 90 ] ) have pushed the limit of simulations to millions (sometimes even billions) of atoms (to include explicit solvent and/or membrane environment) for dynamics of microseconds of time. While we are able to study the spatial and temporal scale of some biological phenomena, simulating membrane curvatures induced by peripheral proteins (such as BAR proteins) is one of the best examples of ultimate applications of molecular dynamics simulations, that is, examining real-time dynamics in addition to sampling the configuration space.
Several members of the BAR superfamily have been extensively studied using computational approaches. We will include the study of N-BAR domains of two proteins, namely, amphiphysin (Section 4.1) and endophilin (Section 4.2); the only molecular dynamics simulation study of the EFC/F-BAR domain (Section 4.3); and the most recent studies of BAR domain with an adjacent PH domain from ACAP1 protein (Section 4.4). We note that, while considerable numerous atomistic information available aids our understanding of the membrane remodeling mechanisms, it also introduces more room for exploration, with unexpected complexity leading to a requirement for further enhancement in methodology.
As a member of the BAR superfamily, N-BAR dimerizes to form a concave surface with positively charged residues (Fig.
One of the aspects that need to be verified for computational studies of membrane sculpting will be whether the simulations could reproduce the membrane curvatures observed in experiments. However, in vivo , membrane curvatures are actually induced by collective motions of membrane remodeling proteins. [ 35 ] While all-atom MD simulations are too computationally costly to simulate lattices of proteins on the membrane, Arkhipov et al. [ 93 ] presented coarse-grained (CG) MD simulations of protein lattices on the membrane, producing membrane curvatures comparable to the experimental results. While altering the arrangement of proteins inside the protein lattices could result in a maximum of four-fold effect on the induced membrane curvature, it addressed one of the uncertainties of the CG models, that is, the atomistic information on interactions between proteins inside the lattice was crucial but unclear. Yin et al. [ 56 ] presented even larger simulations of up to 43 proteins on membranes by using their shape-based coarse-grained (SBCG) models and achieved closing of curved membranes with two ends. A number of speculations have been made on the basis of arrangements of proteins inside the protein lattices producing the membrane curvatures closest to that measured in experiments, but atomic-level evidence was insufficient. CG MD simulations also advanced the understanding of the role of the N-terminal helices in inducing membrane curvatures. Directly removing the amphipathic helices of 8 N-BAR domains in CG MD simulations still generated considerable membrane curvature. [ 57 ] In addition to the observation that simulations with helices alone fail to recapture any remodeled membrane, Arkhipov et al. [ 57 ] concluded that scaffolding is the key mechanism by which N-BAR domains bend lipid bilayers. However, the fact that the all-atom MD simulations conducted by Blood et al. [ 92 ] show membrane curvatures induced solely by amphipathic helices keeps room for debate on the actual role of the N-terminal helix in the N-BAR domain.
A few groups of researchers have studied the membrane-remodeling mechanism of N-BAR domains through simulations of proteins and lipids in water. Ayton et al. [ 94 ] constructed and refined [ 95 ] a mesoscopic model which considers the effective curvature field replacing individual proteins to reproduce the membrane remodeling process (Fig.
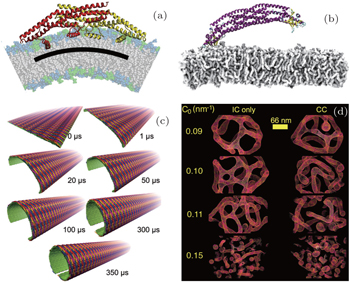 | Fig. 3. (a) All-atom MD simulations by Cui et al . [ 97 ] demonstrating symmetrical binding of the N-BAR domain of endophilin protein. Reproduced with permission from Ref. [ 97 ]. Copyright by 2009 Biophysical Society. (b) All-atom MD simulations by Pang et al . [ 80 ] capturing asymmetrical binding of BAR-PH domain protein on PIP 2 -containing membrane. (c) CG MD simulations done by Yu et al . [ 58 ] show the dynamic process of closing of the membrane by an array of F-BAR proteins. Reproduced with permission from Ref. [ 58 ]. Copyright 2013 Yu, Schulten. (d) Mesoscopic model developed by Ayton et al . [ 95 ] in turn succeeded in showing tubulation and vesiculation. Reproduced with permission from Ref. [ 95 ]. Copyright by 2009 Biophysical Society. |
The structure of the endophilin N-BAR domain is characterized by two additional amphipathic helices (one on each monomer) located under the top of the concave surface of the N-BAR domain dimer (Fig.
Following a similar line of reasoning in the case of the amphiphysin N-BAR domain, although electrostatic interaction between the concave surface of the protein and negatively charged lipids is believed to dominate in membrane curvature induction, the atomistic details of the amphipathic helices, i.e., how they are inserted into the lipid bilayers and how they affect the arrangement of the proteins inside the protein lattice, may indeed be crucial for determining why the resultant membrane curvature induced by members of the N-BAR sub-family differs for different members. Cui et al. [ 97 ] reported membrane curvatures induced by endophilin N-BAR domains with different orientations of the additional helices using all-atom MD simulations (Fig.
Among the members of the BAR superfamily, members of the F-BAR sub-family (Fig.
While the membrane-sculpting ability of the BAR domains has been fully recaptured both in vitro [ 82 , 101 , 102 ] and in cells [ 72 , 103 ] and is well supported by MD simulations, [ 56 , 58 ] BAR domains from the protein ACAP1 (Fig.
In fact, the PH domain found in GRP1 protein has been previously discussed by Lumb et al . [ 104 ] with respect to its membrane penetrating ability. All-atom MD simulations have been employed to study the characteristics of several binding pockets of the domain and steered MD (SMD) simulations were used to pull the domain away from the membrane in order to study the energy contribution from the different components of the system to the membrane binding stability. The results have shown that the three putative membrane-binding loops of the PH domain are all able to penetrate the lipid bilayer to a variable degree. Among the three loops in the PH domain, one of them (loop β 6/ β 7: residues 340–353) retained interactions with the bilayer for a longer period relative to the other two loops during the SMD simulations, suggesting that this loop contributes to a more pronounced interaction.
Lai et al. [ 105 ] used all-atom MD simulations to study the molecular mechanism underlying membrane binding of the GRP1 PH domain. Their results revealed the full picture of the whole binding process of a PH domain with PIP3 lipids in the membrane. Atomic-scale insights highlighted the PS lipids in the membrane, which attract and orient the PH domain in a long range through electrostatic interactions. This interaction reduced the dimensionality of the search process and was believed to increase the residence time of the PH domain on the membrane surface after binding. Longer residence time might confer the PH domain an improved probability of binding to the rare PIP3 lipids that finally stabilize the binding process. However, in contrast to the conclusion drawn by Lumb et al ., [ 104 ] Lai et al . [ 105 ] observed the largest number of contacts between the phosphate head of the lipid and the loop β 1/ β 2 (residues 275–283).
Recently, Pang et al. [ 80 ] reported that an interesting PH domain found in ACAP1 protein adjacent to the BAR domain induced membrane curvatures. While BAR domains are well known for inducing membrane curvature either through scaffolding (represented by F-BAR domains) or helix insertion (represented by N-BAR domains), the BAR-PH domain of ACAP1 seems to rely upon the PH domains for its membrane-binding ability. The BAR-PH dimers were observed to bind the PIP2-containing membrane asymmetrically, i.e., only one of the two identical monomers binds its PH domain to the membrane while the other binds to the adjacent protein. The asymmetrical geometry was not only observed in experiments but was also successfully recaptured in all-atom MD simulations (Fig.
All-atom MD simulation is one of the well-established methods for studying the biomolecular world. To understand membrane curvature induction by peripheral proteins, atomistic information is of prime importance. First, calculation of the amount of membrane curvature induced by the proteins has been presented by a multitude of studies on different BAR proteins. [ 56 , 58 , 91 – 93 ] These results are of particular importance as membrane curvature is one of the experimental parameters for which one can obtain reliable agreement. Another valuable atomistic information is the stability of certain configurations of the system. For instance, the orientation of amphipathic helices or arrangements of proteins inside a protein lattice have been tested intensively using both all-atom MD simulations and CG MD simulations (to be discussed below). By analyzing some characteristic properties of the system, for example the membrane curvature, we can summarize the effect of the given configuration. Last, with all-atom MD simulations, one can quantitatively evaluate the contributions from different interactions between components of the system. [ 104 ]
A considerable disadvantage of all-atom MD simulation is its limitation at the spatial and temporal scales. While physiological events sometimes take even seconds, it is very difficult for all-atom MD simulation to repeat a completed membrane remodeling process. CG MD simulations therefore appear to fill this vacancy. SBCG MD simulations that extended from atomistic models successfully recaptured closing of a planar membrane sheet into a membrane tube by both N-BAR and F-BAR domain lattices. [ 56 , 58 ] CG MD simulation is proven to be reliable in exploring relatively large scale cellular events and provides membrane tube radii comparable to those obtained experimentally. Only when such large-scale CG MD simulations were used, the effect of arrangement of proteins inside a protein lattice toward the induced membrane curvatures could be addressed. Introducing a number of different arrangements and comparing the resulting curvature with the experimental results has become a common practice to find the most probable configuration. [ 55 , 56 , 58 , 93 ] Here, we note that parametrization of the CG model with a strong physical basis is of prime importance and must be handled with care. The SBCG model uses a self-organizing neural network, which adapts to the shape of the molecule to be represented, to place the CG beads. [ 106 ] The beads inherit the mass and charge of the groups of atoms they represent. Parametrization of interactions between beads bases on information from all-atom simulations or available experimental data. Usually, neighboring beads are connected by harmonic springs and beads from separate molecules interact through 6–12 Lennard–Jones and non-bonded Coulomb potentials. [ 106 , 107 ] The most recent CG MD simulations, which utilized coarse-graining strategies other than SBCG and that performed by Simunovic et al. , [ 108 ] were able to show linear aggregation of N-BAR proteins before membrane remodeling took place. In this study, CG beads are defined based on essential dynamics analyzed from all-atom simulations. [ 109 , 110 ] Sophisticated parametrization of inter-bead interactions in lipids calculates forces from atomic-resolution simulations followed by averaging in a variational manner and supplementing with analytical functions to cover poorly sampled regions of the configurational space. [ 111 – 114 ] For protein beads, a heterogeneous elastic network is assumed and the spring constants are calculated by fitting to thermal averages such as fluctuations of CG pair distances computed from all-atom MD simulations. [ 115 ] While understanding the membrane remodeling process is likely to require information beyond the atomistic details of local curvature induction, CG MD simulations have been shown to be useful in recapturing the interplay between a large numbers of proteins.
Sometimes CG MD simulation is still too computationally costly to be used for examining a huge membrane curvature induction event. A mesoscopic model has been developed and further refined to meet this purpose. [ 94 , 95 , 116 , 117 ] The mesoscopic model only included membranes and an effective curvature field representing the effects of proteins. The mesoscopic simulations were able to reach microseconds and successfully show tubulation of liposomes (Fig.
We note that among the members of the BAR superfamily that have well-resolved crystallography structures, PX-BAR and I-BAR domain proteins have not yet been studied using MD simulations despite their close relation with membrane remodeling processes, [ 3 , 81 – 83 , 119 – 121 ] The PX-BAR domain protein, which has an adjacent PX domain that is similar to the PH domain in the BAR-PH domain protein (Fig.
Although members of the BAR superfamily has been proven to be closely associated with membrane curvatures during many cellular processes, whether they really sculpt or insert helices into membranes and solely provide all the bending energy is still under debate. Recently, Dannhauser et al. [ 122 ] showed that clathrin, which lacks BAR domains and is thought to coat the membrane pit, is in fact powerful enough to scaffold the lipid bilayers through oligomerization, while Stachowiak et al. [ 52 ] showed that protein–protein crowding, with only a tether connecting between proteins and the membrane, can also induce membrane curvature.
Rapidly emerging evidence of the involvement of BAR domain proteins and other membrane-remodeling proteins in various cellular processes, ranging from organelle biogenesis to inter-cell communication, has brought membrane remodeling to the forefront of life science research. Dramatic improvements have been seen in research on curvature and its generation in the past few years with respect to the structure determination of membrane-bound scaffolds and computational model development, which allow systematic study of membrane remodeling mechanisms. Combination of direct visualisation of membrane-bound BAR scaffolds and state-of-art molecular dynamics simulations has revealed a number of unexpected aspects of BAR biology, including asymmetrical binding by an adjacent PH domain [ 80 ] and ability to access both curved and flat bilayers. [ 54 , 101 ] The original view that the intrinsic curvature decides the spatial and temporal characteristics of BAR superfamily members might be over-simplified. The fact that action and recruitment of BAR proteins are subject to much more complex and poorly understood collective protein modes or regulatory mechanisms thus require study with more intensive combination of experimental and computational outputs. With the continuous advances in methodology, future efforts will be directed toward resolving the structure and sequence of events of membrane-bound multi-protein scaffolds and understanding the mechanics behind the scaffolds. Given that the human proteome contains more than 60 members of the BAR superfamily, decoding the membrane curvature generation will certainly mark an unprecedented breakthrough in modern physiology.
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 | |
| 9 | |
| 10 | |
| 11 | |
| 12 | |
| 13 | |
| 14 | |
| 15 | |
| 16 | |
| 17 | |
| 18 | |
| 19 | |
| 20 | |
| 21 | |
| 22 | |
| 23 | |
| 24 | |
| 25 | |
| 26 | |
| 27 | |
| 28 | |
| 29 | |
| 30 | |
| 31 | |
| 32 | |
| 33 | |
| 34 | |
| 35 | |
| 36 | |
| 37 | |
| 38 | |
| 39 | |
| 40 | |
| 41 | |
| 42 | |
| 43 | |
| 44 | |
| 45 | |
| 46 | |
| 47 | |
| 48 | |
| 49 | |
| 50 | |
| 51 | |
| 52 | |
| 53 | |
| 54 | |
| 55 | |
| 56 | |
| 57 | |
| 58 | |
| 59 | |
| 60 | |
| 61 | |
| 62 | |
| 63 | |
| 64 | |
| 65 | |
| 66 | |
| 67 | |
| 68 | |
| 69 | |
| 70 | |
| 71 | |
| 72 | |
| 73 | |
| 74 | |
| 75 | |
| 76 | |
| 77 | |
| 78 | |
| 79 | |
| 80 | |
| 81 | |
| 82 | |
| 83 | |
| 84 | |
| 85 | |
| 86 | |
| 87 | |
| 88 | |
| 89 | |
| 90 | |
| 91 | |
| 92 | |
| 93 | |
| 94 | |
| 95 | |
| 96 | |
| 97 | |
| 98 | |
| 99 | |
| 100 | |
| 101 | |
| 102 | |
| 103 | |
| 104 | |
| 105 | |
| 106 | |
| 107 | |
| 108 | |
| 109 | |
| 110 | |
| 111 | |
| 112 | |
| 113 | |
| 114 | |
| 115 | |
| 116 | |
| 117 | |
| 118 | |
| 119 | |
| 120 | |
| 121 | |
| 122 |