Computational investigations on polymerase actions in gene transcription and replication: Combining physical modeling and atomistic simulations
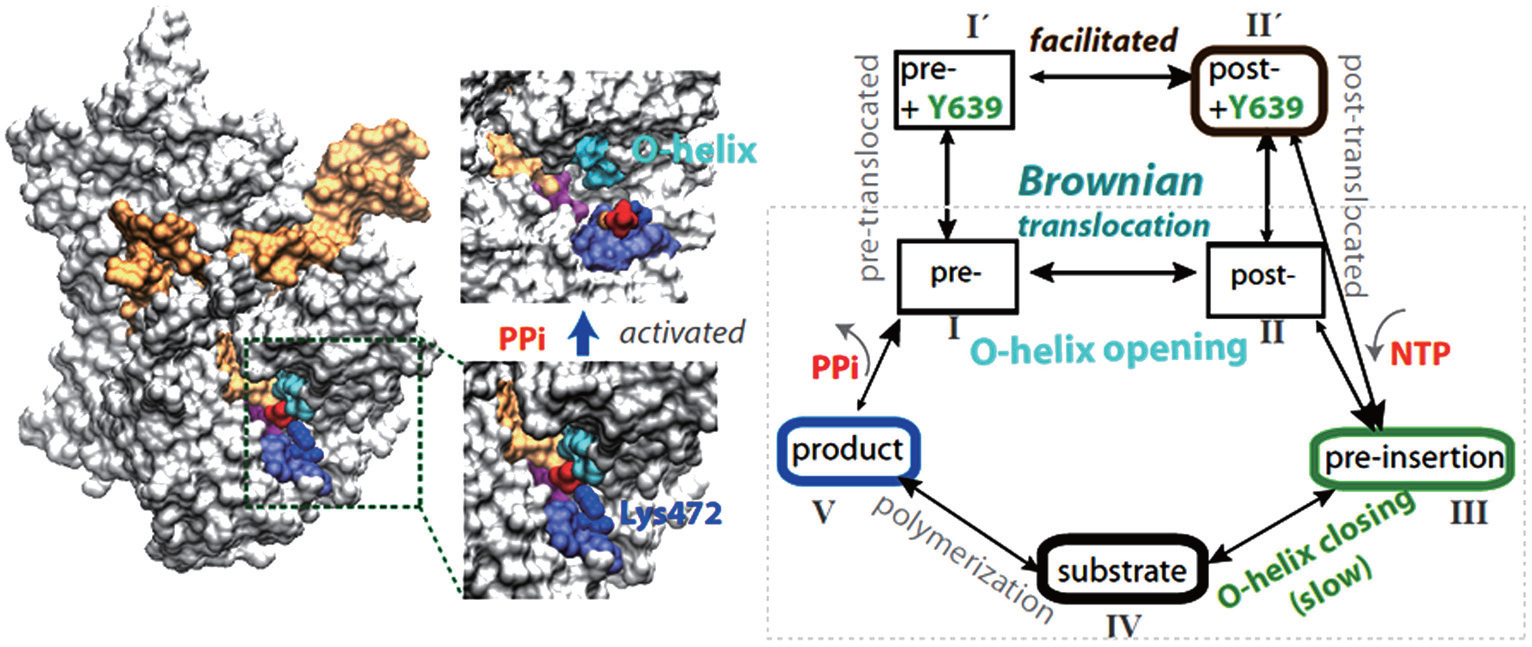
Transcription elongation of T7 RNAP and mechano–chemical coupling features. (a) The product structure of single subunit T7 RNAP elongation complex (in a surface representation: protein, white; NA, orange; the O-helix on the fingers domain, cyan; PPi, red; Lys472, blue, and the linked loop, light blue). The PPi release is found to be a jump-from-cavity process that is assisted by Lys472 side chain swing.[