Uncovering the underlying physical mechanisms of biological systems via quantification of landscape and flux
Free energy landscape quantifications of
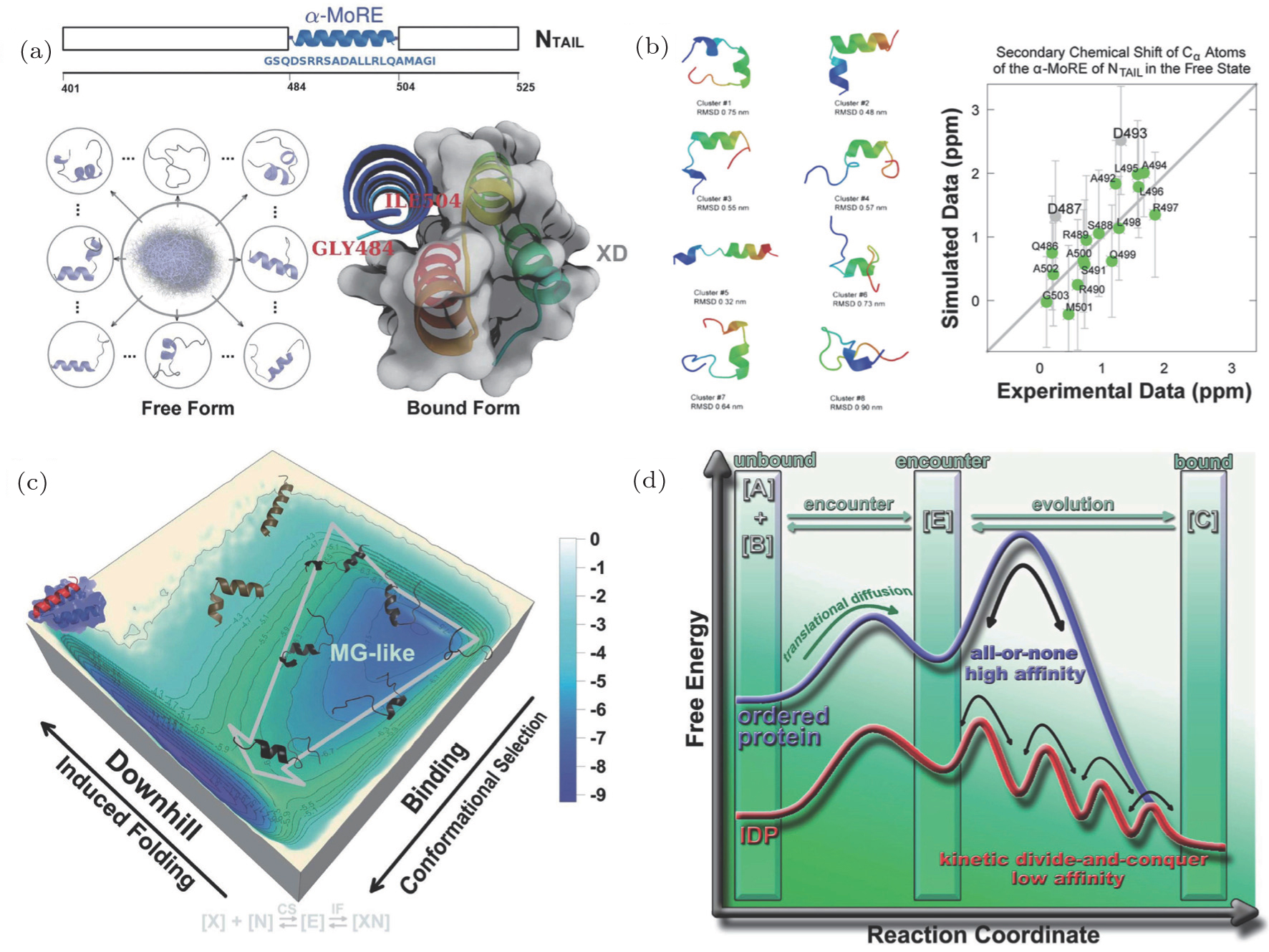
Uncovering the underlying physical mechanisms of biological systems via quantification of landscape and flux |
Free energy landscape quantifications of |
 |