Xu Li 1, †,  , Chu Xiakun 1, Yan Zhiqiang 1, Zheng Xiliang 1, Zhang Kun 1, Zhang Feng 1, Yan Han 1, Wu Wei 1, Wang Jin 1, 2, ‡,  |
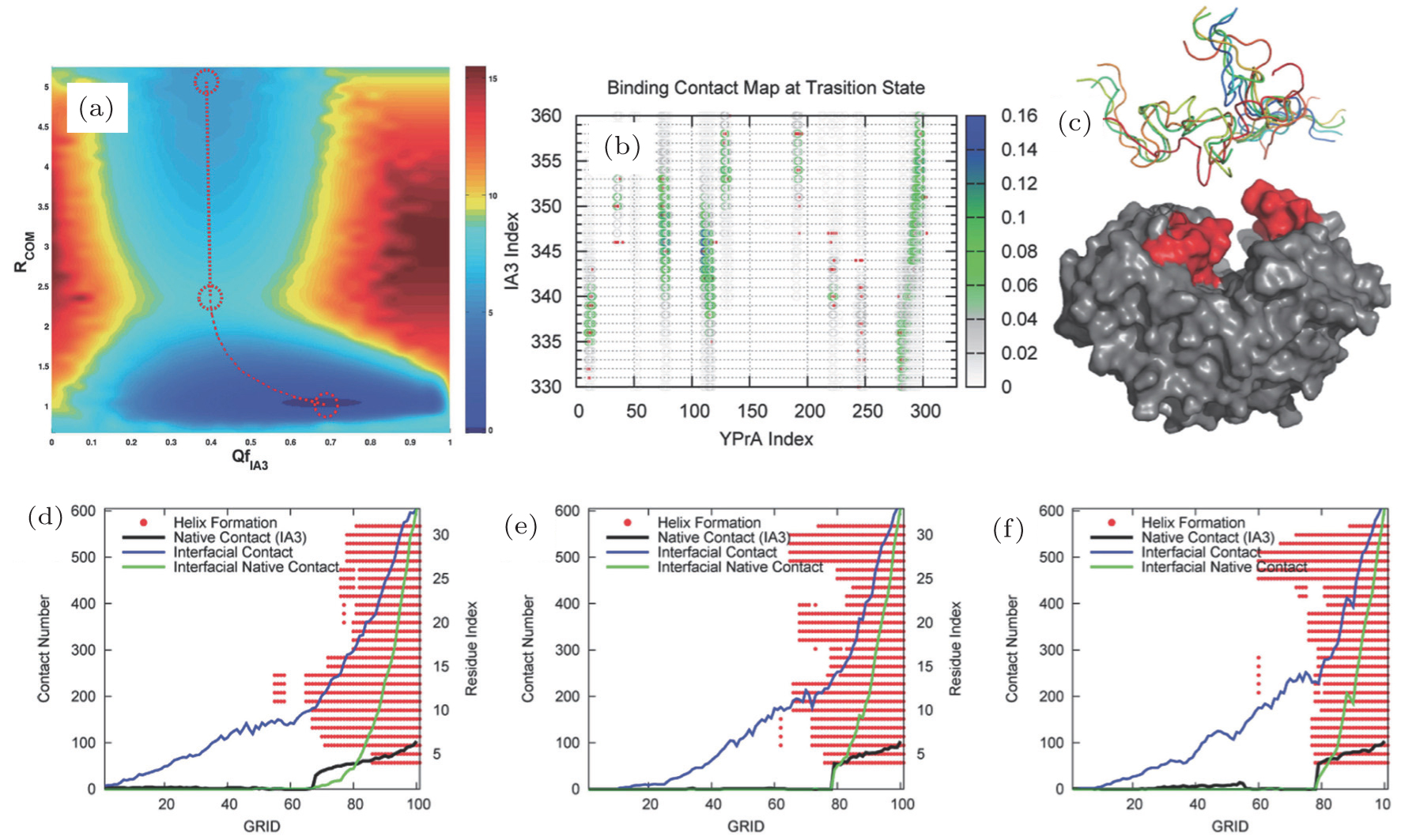
Free energy landscape, contact map, and kinetic pathway analyses of IA3 binding–folding to its target enzyme. (a) Free energy landscape projected onto the reaction coordinates of binding (Rcom, distance between the center of mass of IA3 and the enzyme) and folding (QfIA3, fraction of the native contacts). (b) Contact map at the transition state ensemble. Colored circles represent contacts formed in the transition states and red points indicate native contacts in the final complex. (c) Starting structures in the atomistic path simulation. (d)–(f) Three typical binding–folding pathways for atomistic simulations.[59] |